Team:SJTU-BioX-Shanghai/Project/project1.3
From 2012.igem.org
(Difference between revisions)
(→Membrane Rudder) |
AleAlejandro (Talk | contribs) (→Membrane Rudder) |
||
| Line 25: | Line 25: | ||
__NOTOC__ | __NOTOC__ | ||
<!----------------------------------------------------从这里开始写wiki---------------------------------> | <!----------------------------------------------------从这里开始写wiki---------------------------------> | ||
| - | + | =Membrane Rudder= | |
| - | + | ==Background== | |
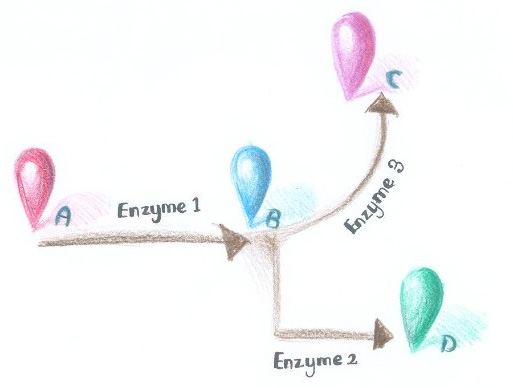
[[Image:12SJTU_divergentreaction.jpg|thumb|270px|right|''Fig.7'' :Demonstration of branched reactions. When signal that induces the dimerization of Enzyme Complex 1 and 2 is present, they would dimerize, making product D more dominant. On the contrary, when signal that induces the dimerization of Enzyme Complex 1 and 3 is present, they would aggregate, so product C is more dominant.]] | [[Image:12SJTU_divergentreaction.jpg|thumb|270px|right|''Fig.7'' :Demonstration of branched reactions. When signal that induces the dimerization of Enzyme Complex 1 and 2 is present, they would dimerize, making product D more dominant. On the contrary, when signal that induces the dimerization of Enzyme Complex 1 and 3 is present, they would aggregate, so product C is more dominant.]] | ||
| Line 42: | Line 42: | ||
<br> | <br> | ||
| - | + | ==Light Signal== | |
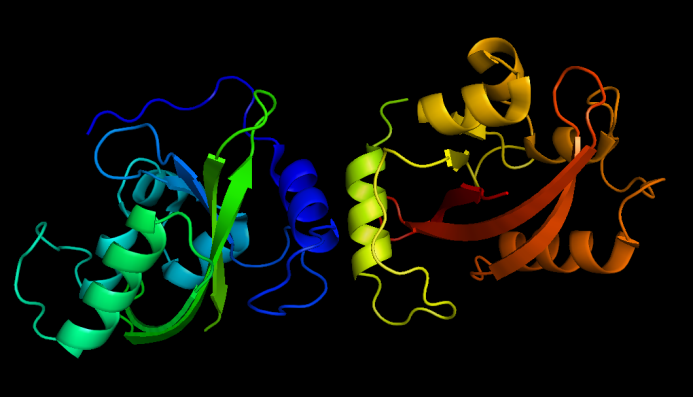
[[Image:VVD dark state.png|thumb|320px|left|''Fig.8'' :Dark state VVD dimers, from PDB ID 2PD7]] | [[Image:VVD dark state.png|thumb|320px|left|''Fig.8'' :Dark state VVD dimers, from PDB ID 2PD7]] | ||
| Line 57: | Line 57: | ||
VVD mutant (C71V and N56K) is harder to dimerize in the dark and easier to disassociate under blue light compared with wildtype VVD. Still, we used split GFP fluorescence complementation assay to test the system. | VVD mutant (C71V and N56K) is harder to dimerize in the dark and easier to disassociate under blue light compared with wildtype VVD. Still, we used split GFP fluorescence complementation assay to test the system. | ||
| - | + | ==Transcription Signal== | |
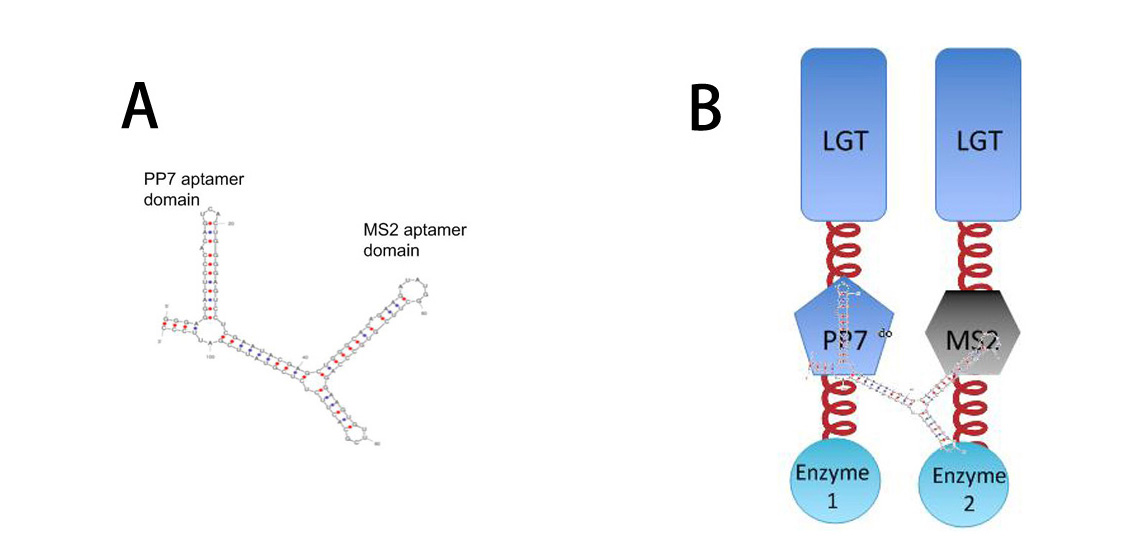
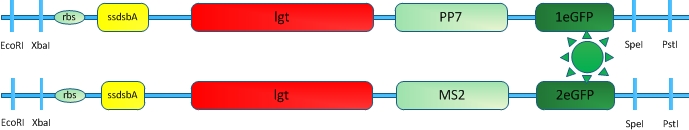
[[Image:12SJTU RNASIGNAL.jpg|thumb|500px|center|''Fig.10'' :A: Sketch of signal RNA molecule which consists of PP7 and MS2 aptamer domains. This RNA molecule is called PM in our system | [[Image:12SJTU RNASIGNAL.jpg|thumb|500px|center|''Fig.10'' :A: Sketch of signal RNA molecule which consists of PP7 and MS2 aptamer domains. This RNA molecule is called PM in our system | ||
| Line 73: | Line 73: | ||
Result | Result | ||
| - | + | ==Chemical Signal== | |
There are many ligand induced dimer in organisms. Chemical signal like glutamate receptor(Kunishima, Shimada et al. 2000), ErbB family are ligand-induced dimers(Lemmon 2009) and estrogen receptor which forms dimer when estrogen is present.(Kumar and Chambon 1988 | There are many ligand induced dimer in organisms. Chemical signal like glutamate receptor(Kunishima, Shimada et al. 2000), ErbB family are ligand-induced dimers(Lemmon 2009) and estrogen receptor which forms dimer when estrogen is present.(Kumar and Chambon 1988 | ||
Revision as of 20:15, 26 September 2012
 "
"