Team:ZJU-China/project.htm
From 2012.igem.org
(Difference between revisions)
| Line 672: | Line 672: | ||
</div><!-- end .acc_container --> | </div><!-- end .acc_container --> | ||
| - | <h2 class="acc_trigger">09 <strong> | + | <h2 class="acc_trigger">09 <strong>PERSPECTIVES</strong></h2> |
<div class="acc_container" style="display: none; "> | <div class="acc_container" style="display: none; "> | ||
<!--Content Goes Here--> | <!--Content Goes Here--> | ||
<div style="height:800px;overflow:scroll;"> | <div style="height:800px;overflow:scroll;"> | ||
| - | <h2>1. | + | <h2>1. Riboscaffold and targeted drug delivery therapy</h2> |
| - | <p | + | <p>This is an extension application of our designed clover series of riboscaffold. Some diseases, such as Cancer, will release some small molecular or change microenvironments beside it thus produce detectable signals. Different from using a biosensor to detect these signals, we utilize our scaffold’s aptamer, accompanying with the production of medicine target the disease. If we change Theophylline aptamer into nidus(disease) molecular aptamer, when riboscaffold bind nidus molecular and change conformation, MS2 aptamer & PP7 aptamer are going to set closer. Enzymes which combining MS2 aptamer & PP7 aptamer and producing drugs are ready to catalyze thus bring out targeting agents. It turns out to be a one-stop agency, once detect the focus of diease, will generate corresponding drug targeting the diease. </p> |
| - | + | <img src=”https://static.igem.org/mediawiki/igem.org/d/da/ZJU_persp_1.png” width=”600px” / > | |
| - | < | + | Figure1. Riboscaffold which can detect and treat diseases. </p> |
| - | <p | + | |
| - | <p | + | <h2>2. Shining Riboscaffold</h2> |
| - | < | + | <p>Paige[1] has reported some RNA aptamers that can bind fluorophores, which are small compounds, and in this way resemble the chemical bonds in GFP, then give out fluorescence. </p> |
| - | <p | + | |
| - | <h2> | + | <p>If we use these aptamer in replace of the theophylline aptamer on our riboscaffold, we can make the riboscaffold shining upon the binding of signal compounds mentioned above. This is a cool method to visualize the states, dynamics and localization of riboscaffold in the living cell. </p> |
| - | <p | + | <img src=” https://static.igem.org/mediawiki/igem.org/9/95/Zju_persp_2.png” width=”600px” / > |
| - | <p | + | <p>Figure2. Aptamers that can shine upon binding. </p> |
| - | < | + | |
| - | < | + | <h2>3. LEGO Riboscaffold</h2> |
| - | < | + | <p>Riboscaffold has unbelievable ability to extend itself through base pairing with each other, just like LEGO bricks! The assembly of LEGO riboscaffolds can load more enzymes and to a large degree accelerate the reaction or artificially construct a longer pathway with high efficiency. For example, artificial TCA cycle abd artificial EMP are promising results. The following pictures show our wide imagination of the possible structure of LEGO riboscaffolds. </p> |
| - | <p | + | |
| - | < | + | <p>But how to obtain these LEGO riboscaffolds? Wachtveitlb[2] has reported a fantastic method to detect RNA-RNA interaction by introducing fluorophores like 1-ethynylpyrene into the 2-position of RNA adenosine. When two single-stranded RNAs with this fluorophore base pair with each other, the fluorescence spectrum changes and thus suggesting their interaction. So it is hopeful to find the desired riboscaffolds as LEGO bricks by selecting from the library! </p> |
| - | <h2> | + | <img src=” https://static.igem.org/mediawiki/igem.org/b/bc/Zju_persp_4.png” width=”600px” / > |
| - | <p | + | <p> LEGO bricks.</p> |
| - | + | <img src=” https://static.igem.org/mediawiki/igem.org/5/56/Zju_persp_5.png” width=”600px” / > | |
| - | + | <p> Long scaffold that has multiple binding sites.</p> | |
| - | <p | + | <img src=” https://static.igem.org/mediawiki/igem.org/e/ea/Zju_persp_6.png” width=”600px” / > |
| - | + | <p> A possible device built by LEGO riboscaffold.</p> | |
| - | <p | + | <img src=” https://static.igem.org/mediawiki/igem.org/8/81/Zju_presp_7.png” width=”600px” / > |
| - | + | <p> Sheets and tubes constructed by LEGO riboscaffolds in vivo.</p> | |
| - | <p | + | |
| - | + | <h2>4. Mimic Long ncRNA</h2> | |
| - | + | <p>In eukaryote, there are naturally produced long non-coding RNAs that attract more and more attention these days and display intriguing potential to act as scaffolds [3]. And our riboscaffold can mimic them and bring their functions to prokaryote. One of the functions is combining related transcription factors and bring them to promoter, as a result enhance the expression of target gene. That is because ncRNA can binds both DNA and Proteins, and can travel freely between nucleus and cytoplasm, which displays great advantage as a bridge. </p> | |
| - | < | + | |
| - | <p | + | <p>Aptamers can be selected in vitro against nearly any target of choice. There are RNA aptamers that can specifically bind some transcriptional regulator. For example, Hunsicker [4] has selected one RNA aptamer that can bind TetR, which usually binds on operator sequence and repress gene expression. So once aptamers mentioned above are designed into a riboscaffold, it can initiate the expression of target genes with higher efficiency. </p> |
| - | + | <img src=” https://static.igem.org/mediawiki/igem.org/1/15/Zju_persp_3.png” width=”600px” / > | |
| - | + | <p>Figure3. Riboscaffold that can bring transcription factors to promoters. </p> | |
| - | + | ||
| - | <p | + | <h2>5. Medicine & Health</h2> |
| - | + | <p>To date, many groups have successfully identified aptamers with a variety of functions, including inhibitory and decoy-like aptamers, regulatable aptamers, multivalent/agonistic aptamers, and aptamers that act as delivery vehicles [5]. </p> | |
| - | + | ||
| - | <p | + | <p>By designing these different aptamers into our RNA scaffold, we can endow our scaffold various potential applications in therapeutics and/or diagnostics. </p> |
| - | <p | + | |
| - | <p | + | <p>For instance, designing inhibitory aptamers that targets VEGF into our RNA scaffold can be used to treat the wet age-related macular degeneration and that has been approved by the FDA in December 2004. </p> |
| + | |||
| + | <p>Designing Decoy-like aptamers that can mimic the target sequce of the proteins into our RNA scaffold can be used as decoys to inhibit binding of transcriptional factors such as HIV-tat, NF-κB, and E2F to their cognate sequences on DNA and thus prevent transcription of target genes and may result in powerful therapeutics for treating many human pathologies. </p> | ||
| + | |||
| + | <p>Designing aptamers behavior as delivery tools into our RNA scaffold can be used to deliver not only some siRNAs to target cells but also toxins, radioisotopes, and chemotherapeutic agents encapsulated in nanoparticles. </p> | ||
| + | |||
| + | |||
| + | <h2>References: </h2> | ||
| + | <p> [1] Jeremy S. Paige, Karen Y. Wu, Samie R. Jaffrey, RNA Mimics of Green Fluorescent Protein science, 2011 vol 333, 642-646. </p> | ||
| + | <p> [2] Josef Wachtveitlb, Joachim W. Engels, ect. RNA as scaffold for pyrene excited complexes, Bioorganic & Medicinal Chemistry 16 (2008) 19-26. </p> | ||
| + | <p> [3] Mitchell Guttman, John L. Rinn. Modular regulatory principles of large non-coding RNAs. Nature. 2012 Feb 15;482(7385):339-46. </p> | ||
| + | <p> [4] Anke Hunsicker, Markus Steber, ect. An RNA Aptamer that Induces Transcription, Chemistry & Biology, 2009,Volume 16, Issue 2, 173–180. </p> | ||
| + | <p> [5] Kristina W. Thiel and Paloma H. Giangrande, Therapeutic Applications of DNA and RNA Aptamers. Oligonucleotides, 2009, Volume 19, Number 3, 209-222. </p> | ||
| + | <p> [6] Thodey, K. & Smolke, C.D. Bringing It Together with RNA. Science 333, 412-413 (2011). </p> | ||
| + | |||
| + | |||
</div> | </div> | ||
</div><!-- end .acc_container --> | </div><!-- end .acc_container --> | ||
Revision as of 19:00, 26 September 2012

 "
"























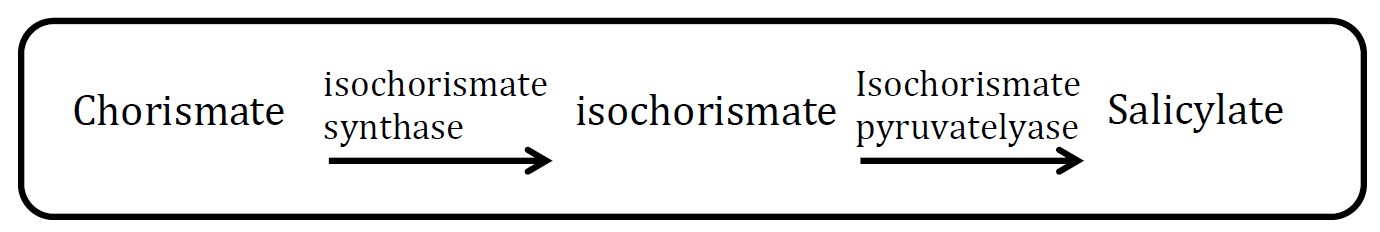









 Figure1. Riboscaffold which can detect and treat diseases.
Figure1. Riboscaffold which can detect and treat diseases.