Team:Cambridge/Project/Biosensors
From 2012.igem.org
| Line 5: | Line 5: | ||
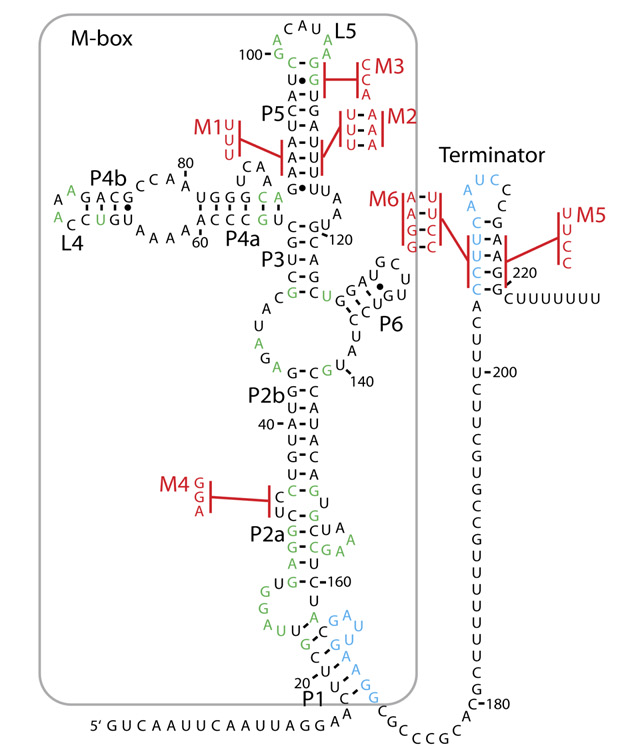
[[File:Magnesium riboswitch image.jpg|350px|right|thumb|The Magnesium Riboswitch we tested. The M-box on the left is the apamer, while the terminator on the right is the expression platform, active only when magnesium is bound. From Dann et al., Cell 2007.]] | [[File:Magnesium riboswitch image.jpg|350px|right|thumb|The Magnesium Riboswitch we tested. The M-box on the left is the apamer, while the terminator on the right is the expression platform, active only when magnesium is bound. From Dann et al., Cell 2007.]] | ||
| - | As our project involved improving the way we make biosensors and apply them in the field, we aimed to develop several biosensors, employing different mechanisms, to prove the extended functionality of the final product. To date, most of the biosensors in the registry use an inducible promoter to control expression of their reporter protein. We aim to use some of these as a proof of the compatability of our kit with a theoretical customers' sensors. However, we also explored another mechanism of biosensing in the form of riboswitches. The rationale behind this decision is found on our <html><a href="https://2012.igem.org/Team:Cambridge/Project/DesignProcess#Ribosens" style="color:#000066">Design process</a></html> page, whilst the success of | + | As our project involved improving the way we make biosensors and apply them in the field, we aimed to develop several biosensors, employing different mechanisms, to prove the extended functionality of the final product. To date, most of the biosensors in the registry use an inducible promoter to control expression of their reporter protein. We aim to use some of these as a proof of the compatability of our kit with a theoretical customers' sensors. However, we also explored another mechanism of biosensing in the form of riboswitches. The rationale behind this decision is found on our <html><a href="https://2012.igem.org/Team:Cambridge/Project/DesignProcess#Ribosens" style="color:#000066">Design process</a></html> page, whilst the success of Ribosense can be found on our <html><a href="https://2012.igem.org/Team:Cambridge/Project/Results#Ribosens" style="color:#000066">Results</a></html> page. |
Riboswitches have several advantages over traditional inducible promotors: firstly, they are usually modular in design, typically containing an aptamer (ligand binding) domain and an expression platform domain, which alters expression of downstream genes. Such a structure may allow for abstraction of discrete parts of each riboswitch, and construction of new regulatory elements with engineered properties that could be designed "de novo" using software tools (Although we may have to wait a little for such modelling capabilities to be realised). Secondly, if the expression platform acts by altering translational rate after transcription has finished, such devices may allow for faster respone times than those seen for current genetic devices, which can only alter transcriptional rates. | Riboswitches have several advantages over traditional inducible promotors: firstly, they are usually modular in design, typically containing an aptamer (ligand binding) domain and an expression platform domain, which alters expression of downstream genes. Such a structure may allow for abstraction of discrete parts of each riboswitch, and construction of new regulatory elements with engineered properties that could be designed "de novo" using software tools (Although we may have to wait a little for such modelling capabilities to be realised). Secondly, if the expression platform acts by altering translational rate after transcription has finished, such devices may allow for faster respone times than those seen for current genetic devices, which can only alter transcriptional rates. | ||
Revision as of 18:49, 26 September 2012


Contents |
Ribosense
As our project involved improving the way we make biosensors and apply them in the field, we aimed to develop several biosensors, employing different mechanisms, to prove the extended functionality of the final product. To date, most of the biosensors in the registry use an inducible promoter to control expression of their reporter protein. We aim to use some of these as a proof of the compatability of our kit with a theoretical customers' sensors. However, we also explored another mechanism of biosensing in the form of riboswitches. The rationale behind this decision is found on our Design process page, whilst the success of Ribosense can be found on our Results page.
Riboswitches have several advantages over traditional inducible promotors: firstly, they are usually modular in design, typically containing an aptamer (ligand binding) domain and an expression platform domain, which alters expression of downstream genes. Such a structure may allow for abstraction of discrete parts of each riboswitch, and construction of new regulatory elements with engineered properties that could be designed "de novo" using software tools (Although we may have to wait a little for such modelling capabilities to be realised). Secondly, if the expression platform acts by altering translational rate after transcription has finished, such devices may allow for faster respone times than those seen for current genetic devices, which can only alter transcriptional rates.
Our sensors
For more information on the biology of each of our parts, please see the links below.
Other Biosensors
We also aimed to use other biosensing parts that were pertinent to the topic of groundwater contamination and which were already available in the registry. Early thoughts of mercury and arsenic testing proved to be unfeasible within the time constraints as our laboratory does not usually handle these materials and so was not already equipped for their use.
Copper [http://partsregistry.org/Part:BBa_K190024 BBa_K190024] and Zinc [http://partsregistry.org/Part:BBa_K190022 BBa_K190022] inducible promoters were selected from the registry, as they seemed promising, with our focus on the copper part as this was labelled as 'working' by the original team Groningen_09. We have sadly been unable to replicate this result through our work over the summer. The capacity of metal ions to induce these promoters is highly dependent on the Heavy metal transporter protein [http://partsregistry.org/Part:BBa_K190018 BBa_K190018], also used by Groningen_09.
One of our initial concerns was that the system may not be compatible in B.subtilis as it was designed in E.coli. Tests in both species were unfavourable however. After several weeks with little success we sent these biobricks to [http://www.lifesciences.sourcebioscience.com/genomic-services/sanger-sequencing-service.aspx Source Bioscience Ltd] for sequencing. While we can now confirm the sequences of the inducible promoters, we found sequencing inconsistencies in the transport protein. This experience has been added to the parts registry.
These inconsistencies seem to have rendered the protein non-functional or sufficiently less functional and time shortages prevented us from looking for an alternative. As such we have been unable to test the capacities of these biobricks in relation to our system.
References
- Nyan Win M. and Smolke C., A modular and extensible RNA-based gene-regulatory platform for engineering cellular function, Proc Natl Acad Sci USA (2007) vol. 104 (36) pp. 14283-8.
 "
"