Team:UC Chile/Results/Int C
From 2012.igem.org
(Difference between revisions)
| Line 13: | Line 13: | ||
</div> | </div> | ||
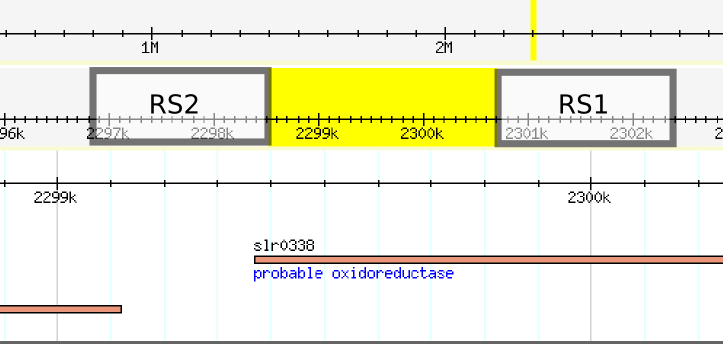
| - | While we did not find any errors in the sequence, we noticed when blasting each separate recombination site, that the position of the recombination sites differed from the ones reported on their page. The position of the sequences of each recombination site are swapped, leaving the backbone part of the plasmid to be integrated into the genome. Furthermore, the recombination sites are separated by aproximately 2.2 kilobases, knocking | + | While we did not find any errors in the sequence, we noticed when blasting each separate recombination site, that the position of the recombination sites differed from the ones reported on their page. The position of the sequences of each recombination site are swapped, leaving the backbone part of the plasmid to be integrated into the genome. Furthermore, the recombination sites are separated by aproximately 2.2 kilobases, knocking out al least one gene. |
<br> | <br> | ||
| Line 26: | Line 26: | ||
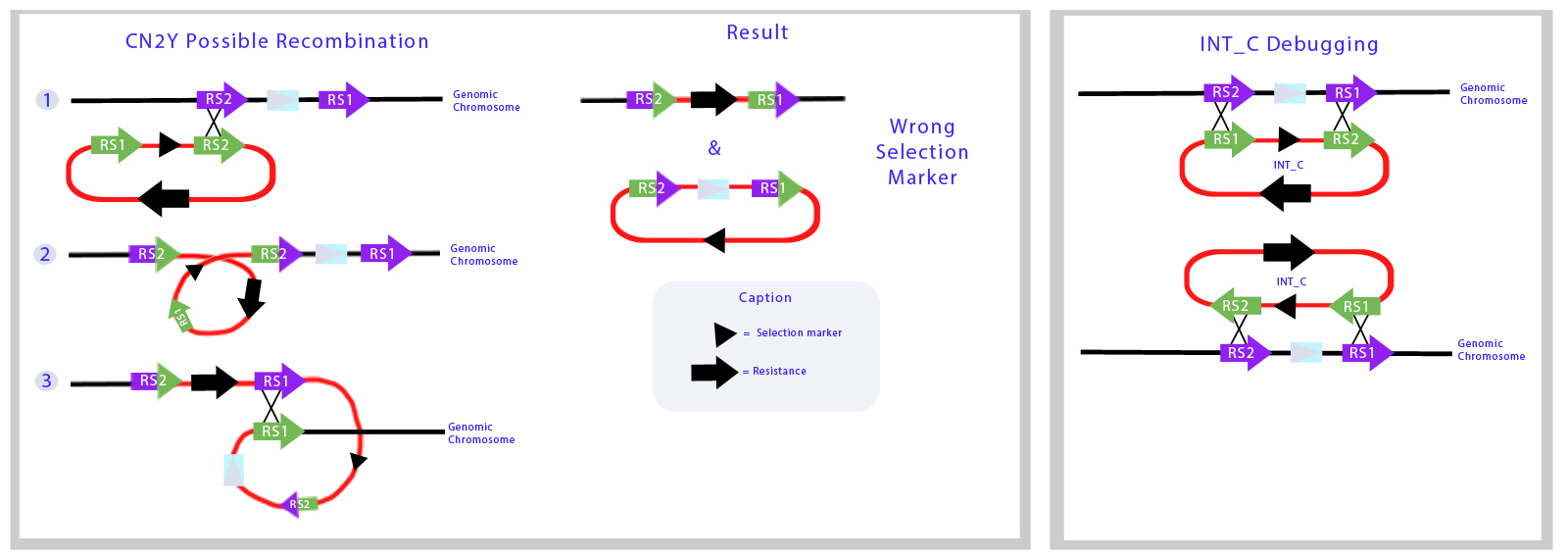
Here we illustrate the recombination mechanism in which the Int_C plasmid would integrate the backbone part of the vector to the genome. | Here we illustrate the recombination mechanism in which the Int_C plasmid would integrate the backbone part of the vector to the genome. | ||
| - | |||
| - | |||
[[File:UC_Chile-CN2Y_Possible_Recombination_Results_and_debugging.jpg|900 px|center]] | [[File:UC_Chile-CN2Y_Possible_Recombination_Results_and_debugging.jpg|900 px|center]] | ||
| + | |||
| + | |||
{{UC_Chilefooter}} | {{UC_Chilefooter}} | ||
Revision as of 18:47, 26 September 2012
 "
"