Team:Cambridge/Project/MagnesiumRiboswitch
From 2012.igem.org
| Line 7: | Line 7: | ||
The riboswitch acts as a transcriptional attenuator when Mg2+ is bound, causing disengagement of the RNA polymerase before it can access downstream ORFs. Consequently, these proteins are not expressed. The system that we used inserted this riboswitch just upstream of the LacI repressor in plasmid pJS130. The lac operator that LacI acted on was upstream of sfGFP, consequently we hope that expression of LacI will lead to repression of sfGFP. | The riboswitch acts as a transcriptional attenuator when Mg2+ is bound, causing disengagement of the RNA polymerase before it can access downstream ORFs. Consequently, these proteins are not expressed. The system that we used inserted this riboswitch just upstream of the LacI repressor in plasmid pJS130. The lac operator that LacI acted on was upstream of sfGFP, consequently we hope that expression of LacI will lead to repression of sfGFP. | ||
| - | [[File:Mg2+construct.png| | + | [[File:Mg2+construct.png|center|700px|thumb|The construct made in the pJS130 vector for the detection of changes in Mg2+ ion concentrations]] |
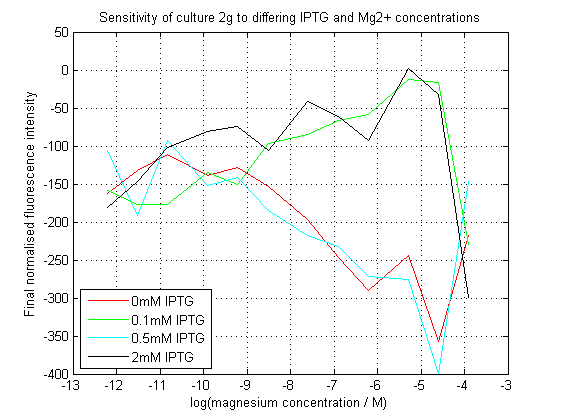
To characterize this construct, we used a 96-well plate reader to assay the effects of the different concentrations of Mg2+ and IPTG on the levels of GFP. We expected the presence of either to allow the expression of sfGFP, however because transcriptional attenuation by the riboswitch occurs before expression of the repressor protein, it was expected that Mg2+ would somehow demonstrate a dominant effect. | To characterize this construct, we used a 96-well plate reader to assay the effects of the different concentrations of Mg2+ and IPTG on the levels of GFP. We expected the presence of either to allow the expression of sfGFP, however because transcriptional attenuation by the riboswitch occurs before expression of the repressor protein, it was expected that Mg2+ would somehow demonstrate a dominant effect. | ||
Revision as of 18:34, 26 September 2012


Magnesium riboswitch
Magnesium is essential for life, being a vital component of many enzymatic reactions. Of particular interest for synthetic biology is its role in the action of DNA polymerase enzymes such as Taq. and Phusion. However, no teams have really characterized a sensor that can be used to measure its concentration in solution. Such a biological sensor exists in the form of the bacillus Mg2+ riboswitch. We attempted to isolate this component and submit it as a biobrick, characterizing its function by inserting it into a derepressor construct.
The riboswitch acts as a transcriptional attenuator when Mg2+ is bound, causing disengagement of the RNA polymerase before it can access downstream ORFs. Consequently, these proteins are not expressed. The system that we used inserted this riboswitch just upstream of the LacI repressor in plasmid pJS130. The lac operator that LacI acted on was upstream of sfGFP, consequently we hope that expression of LacI will lead to repression of sfGFP.
To characterize this construct, we used a 96-well plate reader to assay the effects of the different concentrations of Mg2+ and IPTG on the levels of GFP. We expected the presence of either to allow the expression of sfGFP, however because transcriptional attenuation by the riboswitch occurs before expression of the repressor protein, it was expected that Mg2+ would somehow demonstrate a dominant effect.
We succeeded in creating this construct, and had its sequence verified independently. However, our plate reader assay data was ambiguous - screen-shots of this data can be seen on this page. While a definite trend can be seen relating fluorescence to magnesium concentration, at some IPTG concentrations this trend appears to be inverted compared to others. We are baffled by this result, and hope to repeat the assay to see if we collect similar data.
References
- Dann C., Wakeman C., Sieling C. Baker S. Irnov I. and Winkler W., Structure and Mechanism of a Metal-Sensing Regulatory RNA, Cell 130, 878–892,
 "
"