Team:ZJU-China/project.htm
From 2012.igem.org
(Difference between revisions)
| Line 322: | Line 322: | ||
<div style="height:800px;overflow:scroll;"> | <div style="height:800px;overflow:scroll;"> | ||
<h2>Backround</h2> | <h2>Backround</h2> | ||
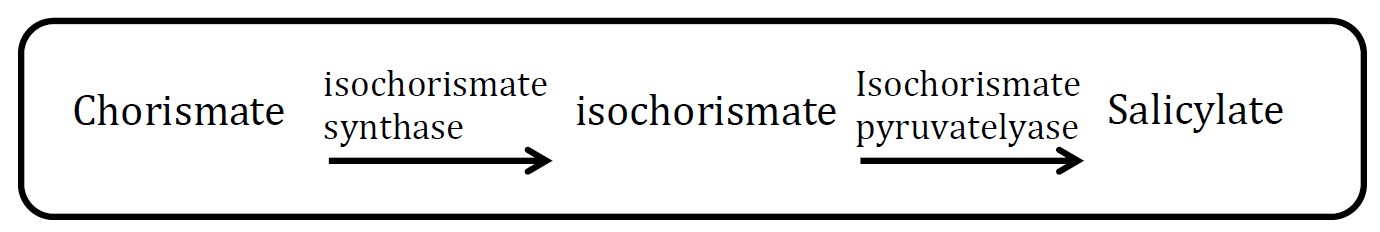
| - | <p>Camille J. Delebecque and his colleagues have designed and assembled RNA structures and used them as scaffolds for the spatial organization of bacterial metabolism ( | + | <p align="justify">Camille J. Delebecque and his colleagues have designed and assembled RNA structures and used them as scaffolds for the spatial organization of bacterial metabolism (Fig.1). Scaffold D0 consists of PP7 and MS2 aptamer domains that bind PP7 and MS2 fusion proteins. As told above, our project is based on the existing scaffold D0. In order to make sure that we can do further work on it, we planned to repeat the work about scaffold D0. </p> |
| - | <p> </p> | + | <p align="justify"> </p> |
<h2>Design</h2> | <h2>Design</h2> | ||
| - | <p> </p> | + | <p align="justify"> </p> |
<img src="https://static.igem.org/mediawiki/2012/d/dc/ZJU_PROJECT_S0_Scaffold_d.jpg" width="600px" /> | <img src="https://static.igem.org/mediawiki/2012/d/dc/ZJU_PROJECT_S0_Scaffold_d.jpg" width="600px" /> | ||
<p> </p> | <p> </p> | ||
| - | <p>Fig.1 How RNA scaffold works. FA and FB represent two halves of EGFP. FA and MS2 are connected with a linker of 30bp. FB and PP7 did the same. The purple scaffold is scaffold D0. MS2 and PP7 can specifically bind to two stem-loops on scaffold, thus | + | <p align="justify">Fig.1 How RNA scaffold works. FA and FB represent two halves of EGFP. FA and MS2 are connected with a linker of 30bp. FB and PP7 did the same. The purple scaffold is scaffold D0. MS2 and PP7 can specifically bind to two stem-loops on scaffold, thus FA and FB get closer and fluoresce under excitation of 480nm.</p> |
<p> </p> | <p> </p> | ||
| - | <h2> | + | <h2>Materials and Methods</h2> |
<p> </p> | <p> </p> | ||
| - | <p> | + | <h3>1. Plasmids and Strains</h3> |
| + | <p align="justify">pCJDFA and pCJDFB respectively comprising the gene of half split EGFP (fragment A and fragment B) and MS2 or PP7 protein were constructed by overlap extension PCR. (See the Overlap PCR protocal) Genes MS2, PP7 and pCJDD0 are provided by Dr. Camille J. Delebecque. pEGFP is provided by Prof. Jianzhong Shao. </p> | ||
<p> </p> | <p> </p> | ||
| - | < | + | <p align="justify">Information of pCJDFA, pCJDFB and pCJDD0 are as the followings:</p> |
| - | + | <h5>1). pCJDFA: FA-MS2 cloned into T7 duet expression vectors pACYCDuet-1 Spr</h5> | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<img src="https://static.igem.org/mediawiki/2012/a/a6/ZJU_PROJECT_S0_PCJDFA.png" width="600px" /> | <img src="https://static.igem.org/mediawiki/2012/a/a6/ZJU_PROJECT_S0_PCJDFA.png" width="600px" /> | ||
<p> </p> | <p> </p> | ||
| - | < | + | <h5>2) pCJDFB (FB-PP7 cloned into T7 duet expression vector pCOLADuet-1) Kanr</h5> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<img src="https://static.igem.org/mediawiki/2012/8/82/ZJU_PROJECT_S0_PCJDFB.png" width="600px" /> | <img src="https://static.igem.org/mediawiki/2012/8/82/ZJU_PROJECT_S0_PCJDFB.png" width="600px" /> | ||
<p> </p> | <p> </p> | ||
| - | < | + | <h5>3) pCJDD0 (Scaffold D0 cloned into T7 duet expression vector PETDuet) Ampr</h5> |
| + | <p><h5>4) BL21-star(DE3)</h5> | ||
| + | <p align="justify">cells were used to co-express plasmids. The most important feature of BL21-star(DE3) is that it carries a mutated rne gene (rne131) which encodes a truncated RNase E enzyme that lacks the ability to degrade mRNA, resulting in an increase in mRNA stability.</p> | ||
| - | < | + | <h3>2. Transformation and induction</h3> |
| - | <p> | + | <p> </p> |
| - | <p> | + | <p align="justify">Three groups of transformation were conducted. The first is BL21-star(DE3) transformed only with pCJDD0, the second with pCJDFA+pCJDFB, and the third with pCJDFA+pCJDFB+pCJDD0. </p> |
| - | <p> | + | <p> </p> |
| - | + | <p align="justify">Pick the single colony to cultivate in 3mL liquid LB with relative resistances. And when OD reached 0.4, induce with 0.2mM IPTG for 2h at 25 degree.</p> | |
| - | < | + | |
<p> </p> | <p> </p> | ||
| - | <p>They were transformed with the pCJDD0 (plasmid with scaffold D0) into BL21-star-(DE3). </p> | + | <p align="justify">Wash the bacteria twice with equivalent PBS. Then test the Fluorescence intensity (FI) and OD with Biotek Synergy Hybrid Reader. |
| + | <p> </p> | ||
| + | <p align="justify">Data was shown in Fig.3. The fluorescence of different expression systems are pictured by Olympus fluoview fv1000 confocal laser scanning microscope ( Fig.2)<p> | ||
| + | |||
| + | |||
| + | <p align="justify">They were transformed with the pCJDD0 (plasmid with scaffold D0) into BL21-star-(DE3). </p> | ||
</div> | </div> | ||
</div><!-- end .acc_container --> | </div><!-- end .acc_container --> | ||
| Line 397: | Line 392: | ||
<div style="height:800px;overflow:scroll;"> | <div style="height:800px;overflow:scroll;"> | ||
| - | <p>Several mutations of RNA scaffold D0 have been designed and made. They show quite different characterizes and functions. With the experiment, more RNA scaffold mutations are characterized. Concept Library of RNA Scaffold is suggested. | + | <p align="justify">Several mutations of RNA scaffold D0 have been designed and made. They show quite different characterizes and functions. With the experiment, more RNA scaffold mutations are characterized. Concept Library of RNA Scaffold is suggested.</p> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<p> </p> | <p> </p> | ||
| - | <p> | + | <p align="justify">What is the Library of RNA Scaffold for? Evolution! The variable of RNA structures accommodates a wide application prospect. Though the point mutation reduced uncertainty of selection and the blindness, trying to find a suitable construction is vast project. Various experimental methods, selection and modeling should be used in this part. By analyzing existing mutations, derivation can be made to construct and find an enhanced RNA scaffold. We called this process evolution. </p> |
<p> </p> | <p> </p> | ||
| - | <p> | + | <p align="justify">The Library may contain changes of self, self-assemble, RNA-RNA interaction, RNA-protein interaction. Some examples are show below.</p> |
<p> </p> | <p> </p> | ||
| - | <p>1. | + | <p align="justify">1. Mutating arm length: changing the arm length of RNA scaffold D0. As the mechanism of D0 is reducing the distance of two key enzyme of the pathway, in other words, the output and reaction efficiency is depend on the local concentration. The two aptamer binding site in our project is on two hairpin arms witch are designed in the same length. The change of the arm length provides feasibility of distance-efficiency research. We used split GFP experiments. We made some mutations with different arm length, the result of D0M4 and D0M 5 split GFP experiment shows the light decreasing lend by split GFP FA-FB distance. The difference (PD0M4=0.079, PD0M5=0.025) suggests that the mutating arm length scaffold doesn’t provide an on/off switch but a definability one. It characterized the D0 in another way.(fig 1:a. D0 is the original scaffold. D0 a-d were mutated to the scaffold with different aptamer arm length. b. The result of arm length mutating. Both D0M4 and D0M5 scaffold half-on GEP.)</p> |
<p> </p> | <p> </p> | ||
| - | <p>Several RNA scaffold mutations are constructed and characterize, but they are the tip of the iceberg. There is still plenty to do in this part. The charms of library are the selection and combination. It introduces a new concept of biobrick combination mode.</p> | + | <p align="justify">1.1 Mutating aptamer binding site: Mutating the PP7 and MS2 binding sites prevented protein scaffolding. Preventing protein scaffolding lead to the key enzyme dissociation and the decrease of enzyme local concentration. By chancing the sequence of MS2 aptamer binding site, the fluorescent light decreased. D0M3 in our project is the molecular with mutated aptamer binding site. Split GFP experiment shows that there is a significant difference between D0 an D0M3(P≦0.05, fig2.c). Camille J. Delebecque has done the same work for the H2 biosynthesis pathway.(fig2: a. MS2 and PP7 bind to the scaffold and make GFP work. b. By mutating aptamer binding site, scaffolding is stop. c. significant difference between D0 an D0M3 )</p> |
| + | <p align="justify"> </p> | ||
| + | <p align="justify">1.2 Assemblage: adding extra sequence for self-, RNA-, protein-assemblage. The added sequence may be a riboswitch, RNA or protein binding site, self-assemble structure. Regulation molecular search is also wanted synchronously. </p> | ||
| + | <p align="justify"> </p> | ||
| + | <p align="justify">Applications and outlook</p> | ||
| + | <p align="justify"> </p> | ||
| + | <p align="justify">1.3 sRNA regulation: Simple an direct RNA-RNA interaction change the object RNA scaffold structure. As a Foundation regulation, it substantially enhances the possibilities of forthcoming experiment. (fig3 The designed scaffold has a interaction to regulatory sRNA. Same mechanism, regulatory molecule can be changed to mRNA a. Turn off the scaffold by the competitive binding with aptamer binding site (green) b. The RNA scaffold has a secondary structural switch controls accessibility of sRNA-binding sites(blue) witch can change the arm length. Output regulated by arm length change. c. both methods were used. d. bind an release the object molecular.)</p> | ||
| + | <p align="justify"> </p> | ||
| + | <p align="justify">1.4 Protein expression (mRNA) regulation: RNA scaffold as a free molecular in cell can specific bind mRNA and protein. Binding molecular changes the structure of scaffold to release or combine something. So that oncogene and virogene can be found and controlled by the drug from RNA scaffold. The problem of cancer therapeutic drug side effecting may solved by it. </p> | ||
| + | <p align="justify"> </p> | ||
| + | <p align="justify">1.5 Self quenching(Self regulation): Adding self binding site, a balance of “on” and “off” scaffolds is built. The relationship between the binding site size, CG bases, binding form and the rate binding molecular is urgently modeled. Forming dimerization and trimerization, the concentration of working scaffold could be regulated.</p> | ||
| + | <p align="justify"> </p> | ||
| + | <p align="justify">1.6 Polo-scaffold: Scaffold with intermolecular binding component. These scaffolds bind each other or bind through mediate molecular. And this binding mode has been proved both in vitro and vivo. The aggregation of molecular also makes artificial organelle achievable. (fig4 a.b. Dimerization and trimerization. Protein binding site is sealed off by the scaffolds themselves. Too much scaffold molecular lend to the self regulation. c. Polo-scaffold be made by head-tail binding and d. mediate molecular binding.)</p> | ||
| + | <p align="justify"> </p> | ||
| + | <p align="justify">Several RNA scaffold mutations are constructed and characterize, but they are the tip of the iceberg. There is still plenty to do in this part. The charms of library are the selection and combination. It introduces a new concept of biobrick combination mode.</p> | ||
Revision as of 14:48, 26 September 2012

 "
"