|
|
| Line 117: |
Line 117: |
| | | | |
| | [[File:Rust4.png|left|450x450pxpx]] [[File: rust5.png|right|450x450pxpx]] | | [[File:Rust4.png|left|450x450pxpx]] [[File: rust5.png|right|450x450pxpx]] |
| - |
| |
| - |
| |
| - |
| |
| - | == Scripting ==
| |
| - |
| |
| - | '''Differential equation solver'''
| |
| - |
| |
| - | '''RALSTONIA'''
| |
| - |
| |
| - | These scripts create the differential equations, find the steady state concetrations and then solve them by a 4th order Runge Kutta:
| |
| - | ----
| |
| - |
| |
| - | '''DIFFERANTIAL EQUATIONS DECLARATIONS'''
| |
| - |
| |
| - | %THIS CODE CREATE ALL THE DIFFERENTIAL EQUATIONS FOR THE SYSTEM FOR %RASTONIA
| |
| - | function y=ecuaDifR(t,v)
| |
| - | %---------Parameters------%
| |
| - | global alfS %Basal concentration of the sensor phcS
| |
| - | global alfRA %Basal concnetration of the comple pchA-pchR
| |
| - | global alfR %Basal concentration of LuxR
| |
| - | global alfI %Basal concentration of CI
| |
| - | global alfCI %Basal concentration of CI
| |
| - | global alfHA %Basal concnetration of HipA7
| |
| - | global alfHB %Basal concnetration of HipB
| |
| - | global alfAS %Basal concnetration of Salycilic acid
| |
| - | global gammaS %Degradation of the sensor pchS
| |
| - | global gammaRA %Degradation of the complex pchR-pchA
| |
| - | global gammaR %Degradation of LuxR
| |
| - | global gammaI %Degradation of LuxI
| |
| - | global gammaCI %Degradation of CI
| |
| - | global gammaHA %Degradation of HipA7
| |
| - | global gammaHB %Degradation of HipB
| |
| - | global gammaAS %Dergradation of Salycilic acid
| |
| - | global mOHS %Kinetic constant for the detection of 3-OH-PAME by the sensor pchS (phosphorylation)
| |
| - | global mSFR %Kinetic constant for the phosphorylation of the complex pchR-pchA by the sensor
| |
| - | global mA %Kinetic constant for the activation of the promoter by the pchA
| |
| - | global mIR %Kinetic constant for the formation of the complex LuxILuxR
| |
| - | global mI %Constant that represent the union of the complex LuxILuxR with the promoter
| |
| - | global mHAHB %Kinetic constant for the inhibition of HipA7
| |
| - | global betaI %Max production of LuxI
| |
| - | global betaCI %Max production of CI
| |
| - | global betaHB %Max peoduction of HipB
| |
| - | global betaHA %Max production of HipA7
| |
| - | global betaAS %Max production of Salicylic acid
| |
| - | global kA %Constant k of the hill ecuation for the promoter promoted by pchA
| |
| - | global kIR %Constant k of the hill equiation for the promorer prmoted by the complex luxIluxR
| |
| - | global kCI %Constant k of the hill equation for the promoter promoted by CI
| |
| - | global hA %Hill constant for the promoters promoted by pchA
| |
| - | global hIR %Hill constant for the promoter promoted by the complex IR
| |
| - | global hCI %Hill constant fot the promoter CI
| |
| - | global eI %Export factor of LuxI
| |
| - | global jI %Import factor of LuxI
| |
| - | global deltaI %Difusion of LuxI outside the cell
| |
| - | global eAS %Export of Salicylic acid
| |
| - | global numcel %number of cells
| |
| - |
| |
| - | if (t<(10) || ((t)>20))
| |
| - |
| |
| - | OH=0;
| |
| - |
| |
| - | else
| |
| - |
| |
| - | OH=15;
| |
| - |
| |
| - |
| |
| - | end
| |
| - |
| |
| - | %------ Variables%------
| |
| - |
| |
| - | S=v(1); %Cocentration of the sensor pchS the cell
| |
| - | SF=v(2); %Concentration of phosphorylated sensor the cell
| |
| - | RA=v(3); %Concentration of the comple pchR-pchA
| |
| - | A=v(4); %Concentratio of the promoter avtivator pchA
| |
| - | Ii=v(5); %Concentration of LuxI inside the cell
| |
| - | Io=v(6); %Concentration of LuI outsied the cell
| |
| - | IR=v(7); %Concentration of the complex LuxI-LuxR
| |
| - | R=v(8);%Concentration of the protein CI
| |
| - | CI=v(9);%Concentration of HipA7
| |
| - | HB=v(10);%Concnetratio of HipB
| |
| - | HA=v(11);%Concentration of salicylic acid
| |
| - | AS=v(12); %Concentratio of quitin monomers
| |
| - |
| |
| - | %---Equations---%
| |
| - |
| |
| - |
| |
| - | dS=alfS- gammaS*S - mOHS*OH*S; %Change of the sensor pchS
| |
| - | dSF = mOHS *OH*S - mSFR *SF*RA ; %Change of phosphorylated sensor
| |
| - | dRA=alfRA - gammaRA*RA - mSFR*SF*RA;%Change of the comple pchR-pchA
| |
| - | dA= mSFR*SF*RA-mA*A; %Change of the activator pchA inside the cell
| |
| - | dIi= alfI+ (betaI*(A^hA))/(kA^hA+(A^hA)) -gammaI*Ii +jI*Io- eI*Ii- mIR*Ii*R; %Change of LuxI inside the cell
| |
| - | dIo= numcel*(eI*Ii-jI*Io)-deltaI*Io; %Change of LuxI outside the cell
| |
| - | dIR= mIR*Ii*R - mI*IR; %Change of the complex LuxI luxR
| |
| - | dR= alfR-gammaR*R -mIR*Ii*R +(betaI*(A^hA))/(kA^hA+(A^hA)); %Change of LuxR
| |
| - | dCI= alfCI -gammaCI*CI+ (betaCI*(CI^hCI))/(kCI^hCI+(CI^hCI)) +(betaCI*(IR^hIR))/(kIR^hIR+(IR^hIR));%Change of CI
| |
| - | dHB=alfHB-gammaHB*HB+(betaHB*(CI^hCI))/(kCI^hCI+(CI^hCI))+(betaHB*(IR^hIR))/(kIR^hIR+(IR^hIR))-mHAHB*HA^2*HB^2; %Chanche of HipB
| |
| - | dHA=alfHA-gammaHA*HA+ (betaHA*(CI^hCI))/(kCI^hCI+(CI^hCI))-mHAHB*HA^2*HB^2; %Change of HipA7
| |
| - | dAS=alfAS-gammaAS*AS +(betaAS*(CI^hCI))/(kCI^hCI+(CI^hCI))-eAS*AS+(betaAS*(IR^hIR))/(kIR^hIR+(IR^hIR)); %Change of Salicylic acid
| |
| - | y1(1)=dS;
| |
| - | y1(2)=dSF;
| |
| - | y1(3)=dRA;
| |
| - | y1(4)=dA;
| |
| - | y1(5)=dIi;
| |
| - | y1(6)=dIo;
| |
| - | y1(7)=dIR;
| |
| - | y1(8)=dR;
| |
| - | y1(9)=dCI;
| |
| - | y1(10)=dHB;
| |
| - | y1(11)=dHA;
| |
| - | y1(12)=dAS;
| |
| - | y=y1';
| |
| - |
| |
| - | end
| |
| - |
| |
| - |
| |
| - |
| |
| - | ----
| |
| - |
| |
| - | '''RUNGE KUTTA'''
| |
| - |
| |
| - | %FILE THAT SOLVES THE DIFFERENTIAL EQUATION AND GRAPHS THEM
| |
| - | alfS=0.9; %Basal concentration of the sensor pchS
| |
| - | alfRA=0.9; %Basal concnetration of the complez pchA-pchR
| |
| - | alfR=0.6; %Basal concentration of LuxR
| |
| - | alfI=0.4; %Basal concentration of LuxI
| |
| - | alfCI=0.5; %Basal concentration of CI
| |
| - | alfHA=1; %Basal concnetration of HipA7
| |
| - | alfHB=0.4; %Basal concnetration of HipB
| |
| - | alfAS=0.4; %Basal concnetration of Salycilic acid
| |
| - | gammaS=1; %Degradation of Chitinase inside the cell
| |
| - | gammaRA=1; %Degradation of chitoporin
| |
| - | gammaR=1; %Degradation of LuxR
| |
| - | gammaI=1; %Degradation of LuxI
| |
| - | gammaCI=1; %Degradation of CI
| |
| - | gammaHA=1; %Degradation of HipA7
| |
| - | gammaHB=4; %Degradation of HipB
| |
| - | gammaAS=1; %Dergradation of Salycilic acid
| |
| - | mOHS=4; %Kinetic constant for the formation of the phoshorilation of the sensor
| |
| - | mSFR=2.6; %Kinetic constant of the reaction of the phosphorilarion of the comple R
| |
| - | mA=3.5; %Kinetic constant for the activation by A
| |
| - | mIR=3; %Kinetic constant for the formation of the complex LuxILuxR
| |
| - | mI=3; %Constant that represent the union of the complex LuxILuxR with the promoter
| |
| - | mHAHB=12; %Kinetic constant for the inhibition of HipA7
| |
| - | betaI=10; %Max production of LuxI
| |
| - | betaCI=9.96; %Max production of CI
| |
| - | betaHB=9.95; %Max production of HipB
| |
| - | betaHA=10; %Max production of HipA7
| |
| - | betaAS=11.2; %Max production of Salicylic acid
| |
| - | kA=0.1; %Constant k of the hill ecuation for the promoter promoted by S
| |
| - | kIR=0.39; %Constant k of the hill equiation for the promorer prmoted by the complex luxIluxR
| |
| - | kCI=0.055; %Cosntant k of the hill equation for the promoter promoted by CI
| |
| - | hA=1.2; %Hill constant for the promoters promoted by S
| |
| - | hIR=3.4; %Hill constant for the promoter promoted by the complex IR
| |
| - | hCI=2.3; %Hill constant fot the promoter CI
| |
| - | eI=0.5; %Export factor of LuxI
| |
| - | jI=0.8; %Import factor of LuxI
| |
| - | deltaI=0.2;%Difusion of LuxI outside the cell
| |
| - | eAS=0.8;
| |
| - | numcel=1; %number of cells
| |
| - | %----%
| |
| - |
| |
| - | h=100; %Tiempo maximo
| |
| - |
| |
| - | m=0.01; %Longitud de paso [s]
| |
| - |
| |
| - | t=0:m:h; %Vector tiempo
| |
| - |
| |
| - | xi=[1,1,1,1,1,1,1,1,1,1,1,1];
| |
| - |
| |
| - | y=fsolve(@CondInR,xi);
| |
| - |
| |
| - | conInd=y;
| |
| - |
| |
| - | l=(0:m:h)'; %Vector de tiempo
| |
| - |
| |
| - | x=zeros(length(l),length(conInd)); %Matriz de variables, en las columnas varia
| |
| - | %la variable y en las filas varia el tiempo
| |
| - |
| |
| - | OH=zeros(1,length(l));
| |
| - |
| |
| - | x(1,:)=conInd;
| |
| - |
| |
| - | for k=1:length(l)-1
| |
| - |
| |
| - | xk=x(k,:); %Captura de la ultima posicion de la matirz, es decir, los
| |
| - | %valores actuales de las variables
| |
| - |
| |
| - | k1=ecuaDifR(l(k),xk); %Primera pendiente del metodo de RK4
| |
| - | k2=ecuaDifR(l(k)+m/2,xk+(m/2*k1)'); %Segunda pendiente del metodo de RK4
| |
| - | k3=ecuaDifR(l(k)+m/2,xk+(m/2*k2)'); %Tercera pendiente del metodo de RK4
| |
| - | k4=ecuaDifR(l(k)+m,xk+(m*k3)'); %Cuarta pendiente del metodo de RK4
| |
| - |
| |
| - | xk1=xk+m/6*(k1+2*k2+2*k3+k4)'; %Calculo de nuevos valores para las
| |
| - | %variables
| |
| - |
| |
| - | %xk1=xk+m*ecuaDif(l(k),xk)'; %Method of Newton
| |
| - |
| |
| - | xk2=zeros(1,length(xk1));
| |
| - |
| |
| - |
| |
| - | for p=1:length(xk1)
| |
| - |
| |
| - | if(xk1(p)<0.00000001)
| |
| - |
| |
| - | xk2(p)=0;
| |
| - | else
| |
| - |
| |
| - | xk2(p)=xk1(p);
| |
| - | end
| |
| - |
| |
| - | end
| |
| - |
| |
| - |
| |
| - | x(k+1,:)=xk2; %Actualizacion del nuevo vector de variables en la matriz
| |
| - |
| |
| - |
| |
| - |
| |
| - | end
| |
| - |
| |
| - | for j=1:length(l)
| |
| - |
| |
| - | if (l(j)<(10) || l(j)>(20))
| |
| - |
| |
| - | OH(j)=0;
| |
| - |
| |
| - | else
| |
| - |
| |
| - | OH(j)=15;
| |
| - |
| |
| - |
| |
| - | end
| |
| - |
| |
| - |
| |
| - | end
| |
| - |
| |
| - | S=x(:,1);
| |
| - | SF=x(:,2);
| |
| - | RA=x(:,3);
| |
| - | A=x(:,4);
| |
| - | Ii=x(:,5);
| |
| - | Io=x(:,6);
| |
| - | IR=x(:,7);
| |
| - | R=x(:,8);
| |
| - | CI=x(:,9);
| |
| - | HB=x(:,10);
| |
| - | HA=x(:,11);
| |
| - | AS=x(:,12);
| |
| - |
| |
| - | figure(1)
| |
| - | plot(l,S,l,SF)
| |
| - | legend('Sensor (pchS)',' Phosporilated pchS')
| |
| - | xlabel('Time')
| |
| - | ylabel('Concetratio (micromolar)')
| |
| - | title('Response of Sensor pchS')
| |
| - | figure(5)
| |
| - | plot(l,RA,l,A)
| |
| - | legend('Complex pchR-pchA','Activator pchA')
| |
| - | xlabel('Time')
| |
| - | ylabel('Concetration (micromolar)')
| |
| - | title('Activator response')
| |
| - | figure(2)
| |
| - | plot (l,R,l,Ii,l,Io,l,IR)
| |
| - | legend('LuxR','LuxI nside the cell','LuxI outside the cell','Complex(Lux-LuxR)')
| |
| - | xlabel('Time')
| |
| - | ylabel('Concetration (micromolar)')
| |
| - | title('LuxI-LuxR system response')
| |
| - | figure(3)
| |
| - | plot (l,HA,l,HB)
| |
| - | legend('Toxin HipA7','Antitoxin HipB')
| |
| - | xlabel('Time')
| |
| - | ylabel('Concetration (micromolar)')
| |
| - | title('Toxin-Antitoxin module')
| |
| - | figure(4)
| |
| - | plot (l,OH,l,AS,l,CI)
| |
| - | legend('3-OH-PAME','Salicylic acid','CI')
| |
| - | xlabel('Time')
| |
| - | ylabel('Concetration (micromolar)')
| |
| - | title('CI and Salicylic Acid response')
| |
| - |
| |
| - |
| |
| - |
| |
| - | ----
| |
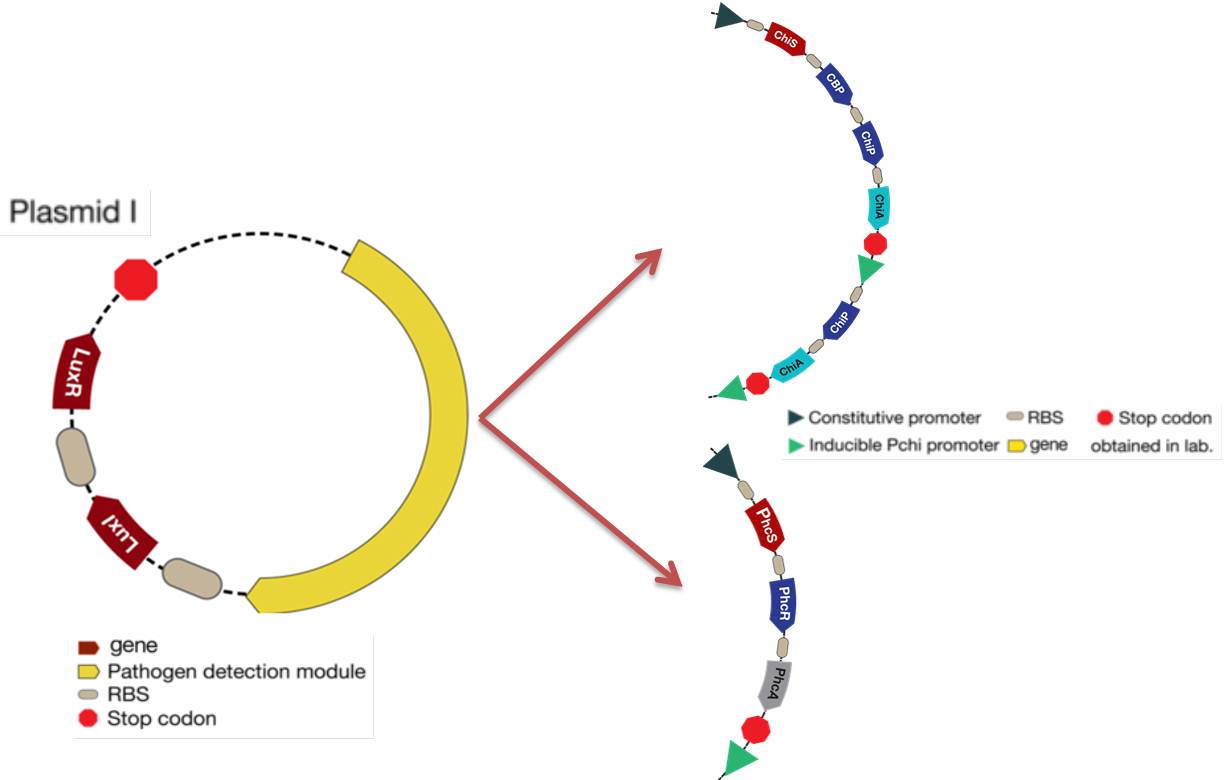
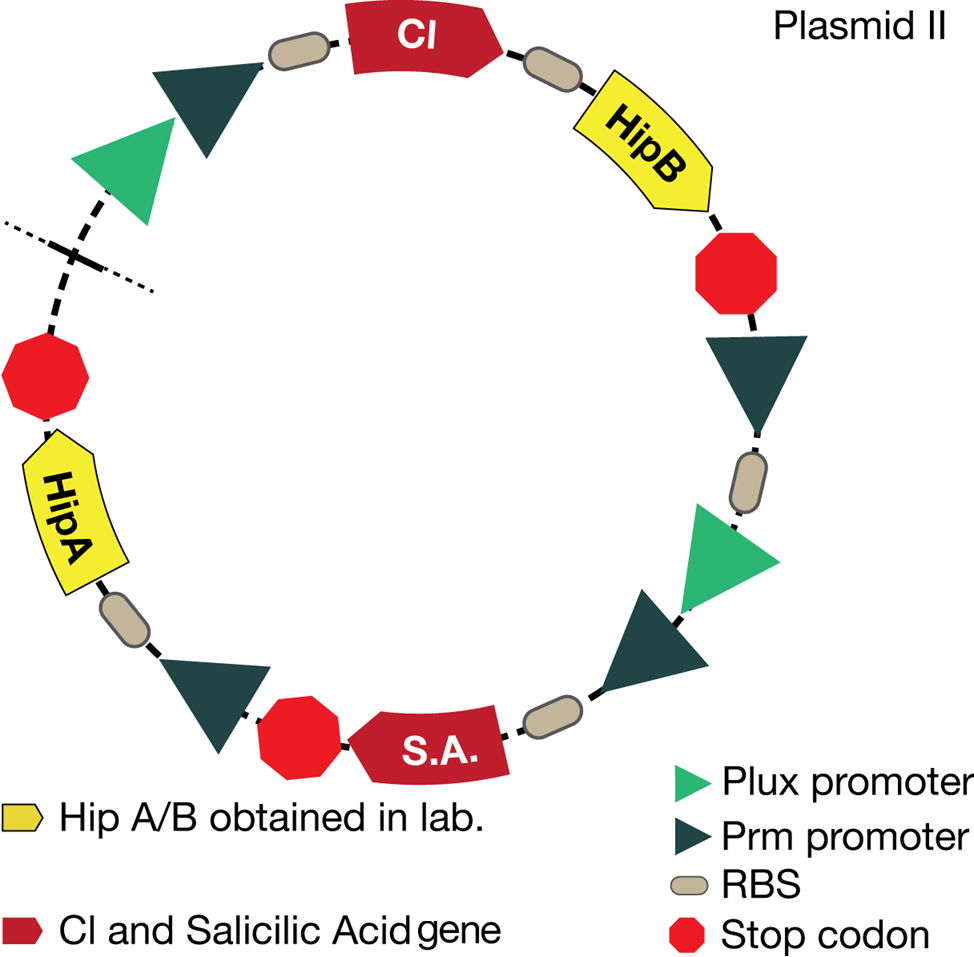
The mathematical model should help the experimental design to optimize the circuit and our case was not the exception. The picture below shows the original design of the circuit.
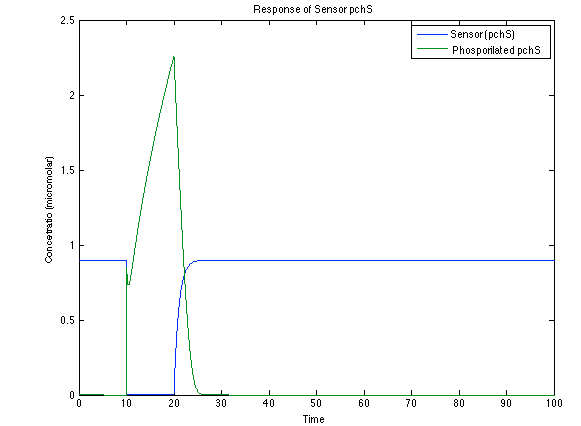
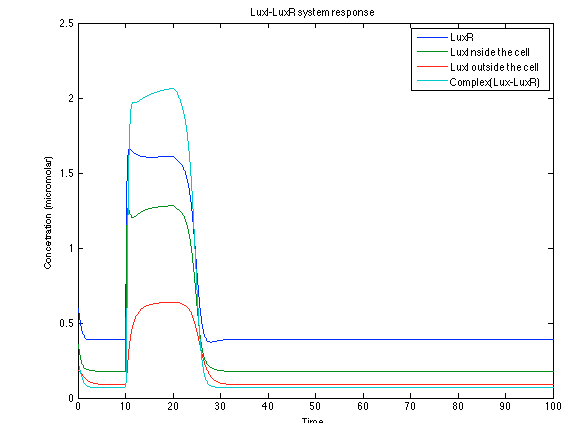
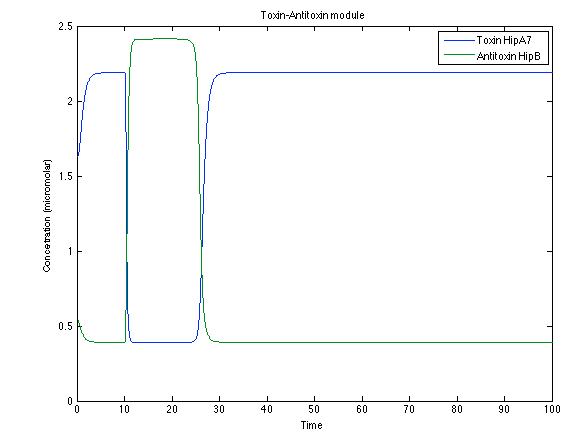
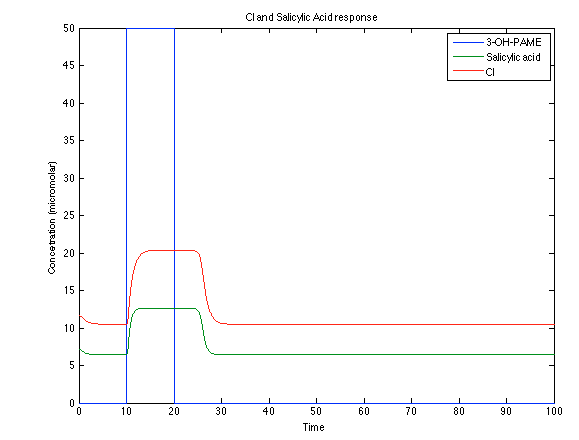
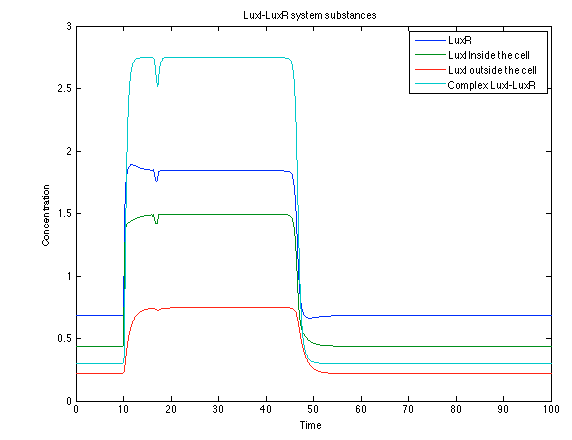
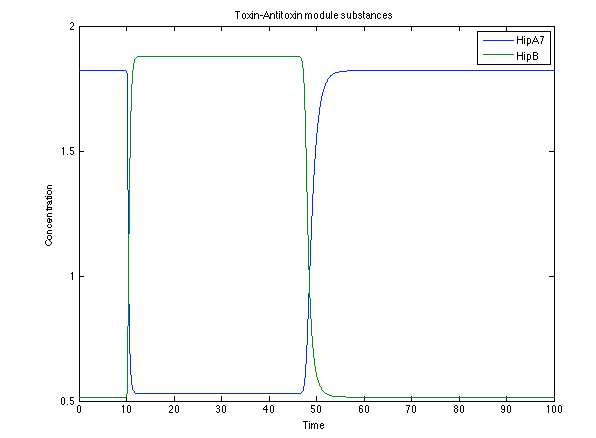
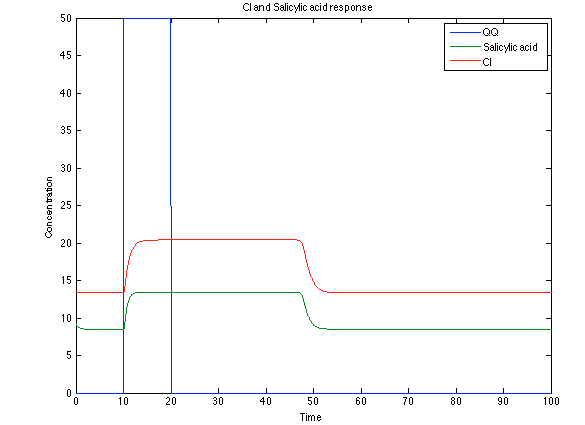
Although the screening of the parameters could not be completely done, we made a manual search for them and found a set that makes the system behave as expected. Here we present the mean response of all the substances in our biological system for one cell. The impulse of the pest was made during the times 10-20.
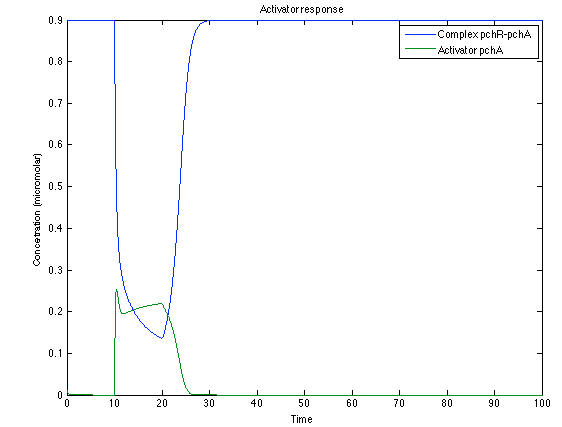
As you see below, when the system is under the presence of 3-OH-PAME the sensor is phosphorylated really fast and the complex phcR-phcA liberates the activator which has a peak and the goes a little down because is with the promoter and its not free. After the impulse is gone everything goes back to normality.
Now, looking at the species of interest, we can see how the antitoxin has greater concentration than the toxin when the 3-OH-PAME appears, this means that the cell is awake and can produce proteins. On the other hand, the Salicylic acid has an increase of almost two fold.
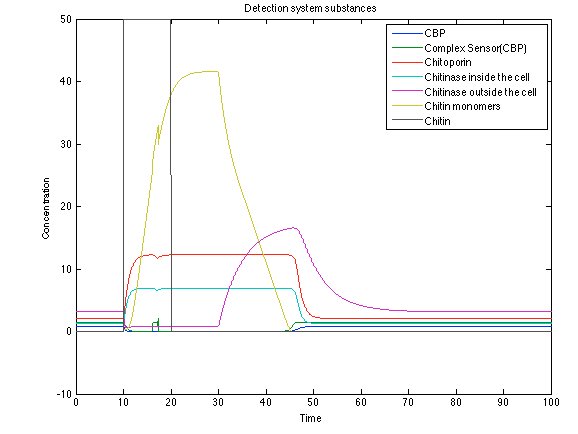
When the system is under the presence of chitin you can see how the positive feedback of the chitoporin and chitinase works and make their concentration increase. This has as a consequence the increase of monomers of chitin and the liberation of the sensor that is gonna activate the LuxI and LuxR promoter.
Like we see with the Ralstonia systema the Lux system's concentrations increase when their promoter is activated. The complex LuxI-LuxR has a peak and then goes down a little because it is busy activation of CI and Salicylic Acid.
The compounds od interest behave just like expected. Like in Ralstonia the cell is awake when the chitin is present and the Salicylic Acid has a two fold increase. It is important to see that this system takes more time to go back to the steady state than the Ralstonia's system.
 "
"