Team:University College London/Module 6/Results
From 2012.igem.org
Sednanalien (Talk | contribs) |
Sednanalien (Talk | contribs) |
||
| Line 19: | Line 19: | ||
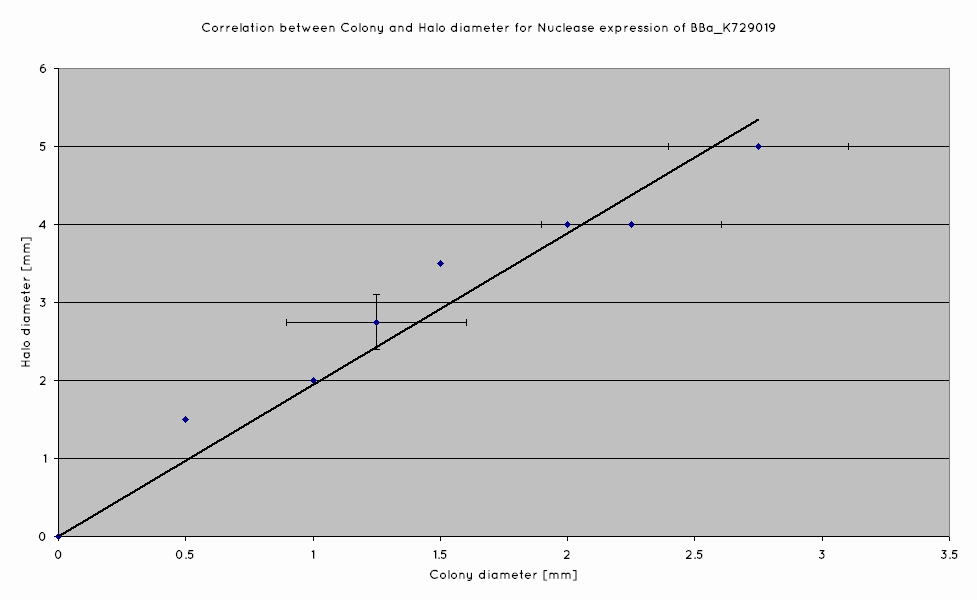
== Colony Halo size Assay == | == Colony Halo size Assay == | ||
| - | + | [[File:UniversityCollegeLondon_Nuclease_Colony_Halo_Diameter_correlation.png]] | |
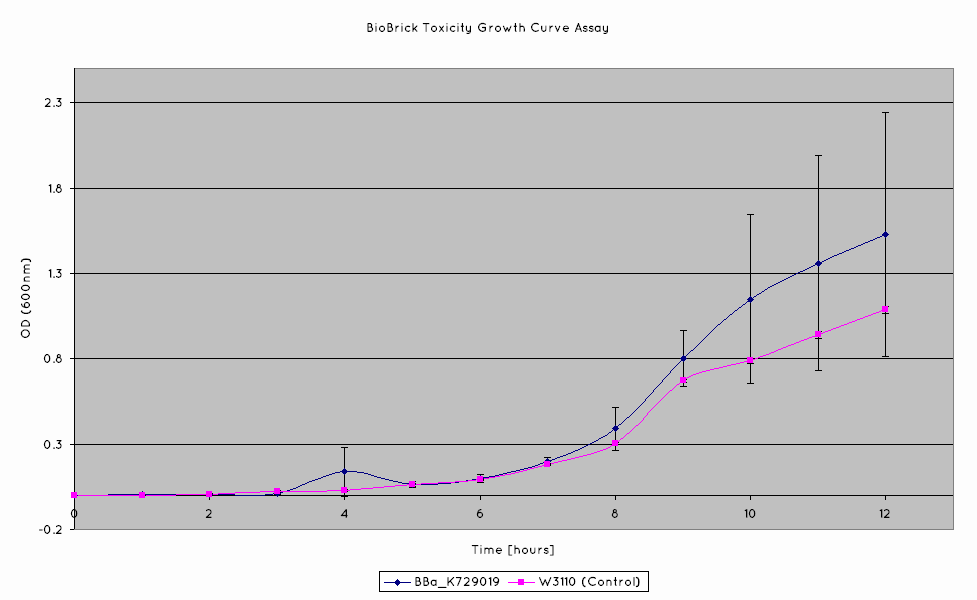
== Nuclease Toxicity Assay == | == Nuclease Toxicity Assay == | ||
| + | |||
| + | [[File:UniversityCollegeLondon_Nuclease_Toxicity_Growth_Curve_Assay.png]] | ||
Revision as of 10:28, 26 September 2012
Contents |
Module 6: Containment
Description | Design | Construction | Characterisation | Modelling | Results | Conclusions
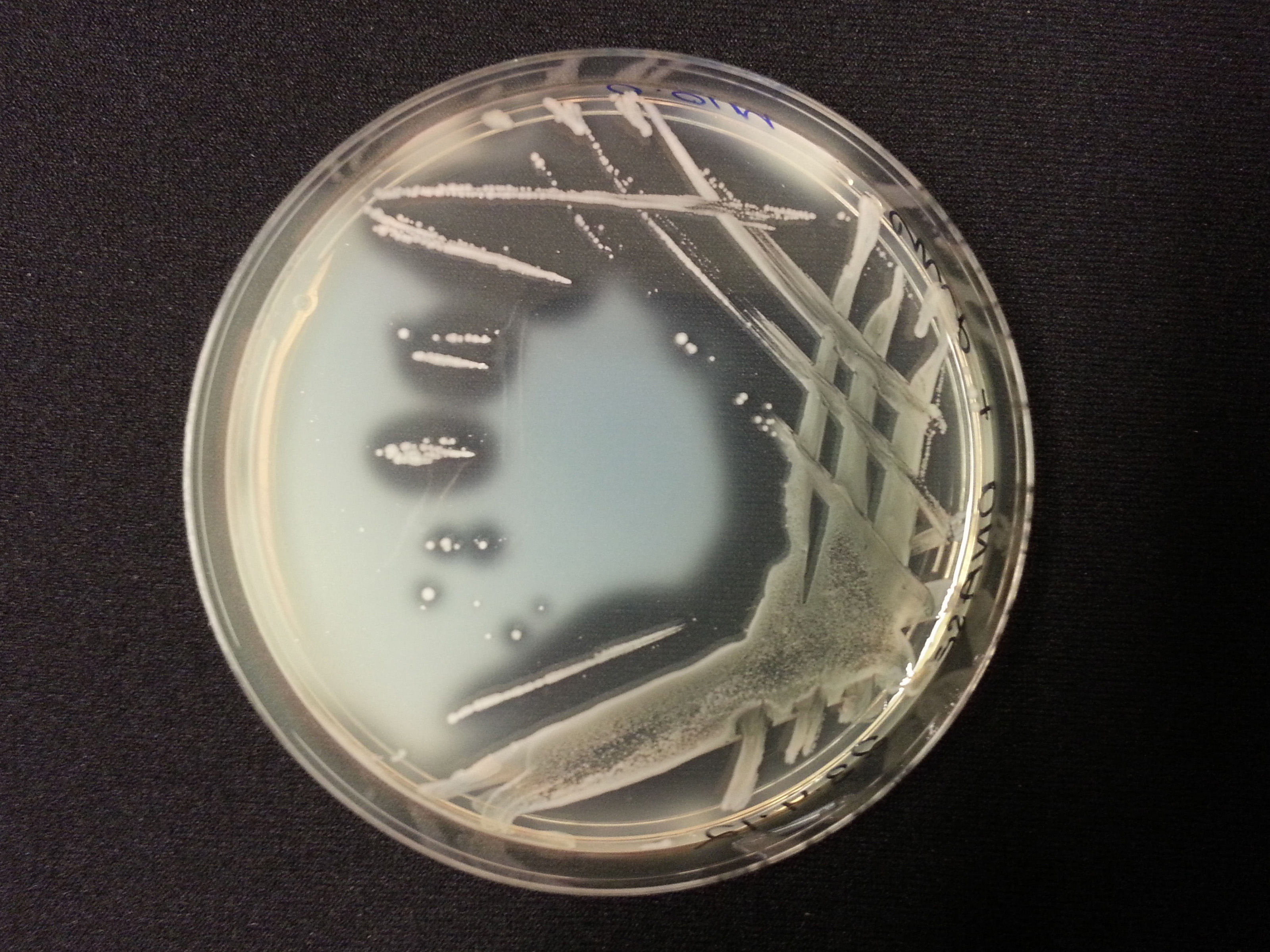
DNase Agar Assay
The following table shows the results we obtained from the DNase agar test. DNase agar contains DNA, which our extracellular nuclease digests. This digestion is observed by adding hydrochloric acid to the agar plates, which causes the remaining DNA to precipitate, causing 'cloudiness' to the DNase agar.
As seen in our DNase agar plate, there is a clear halo surrounding our nuclease transformed cells, indicating a DNA-free zone where the nuclease has digested the DNA in the agar. No such halo is present in our untransformed cells. This indicates that our transformation has been successful, and that BBa_K729004 is working as expected, producing extracellular nuclease that is capable of digesting extracellular genetic material.
| Control | BBa_K729004 |
|---|---|
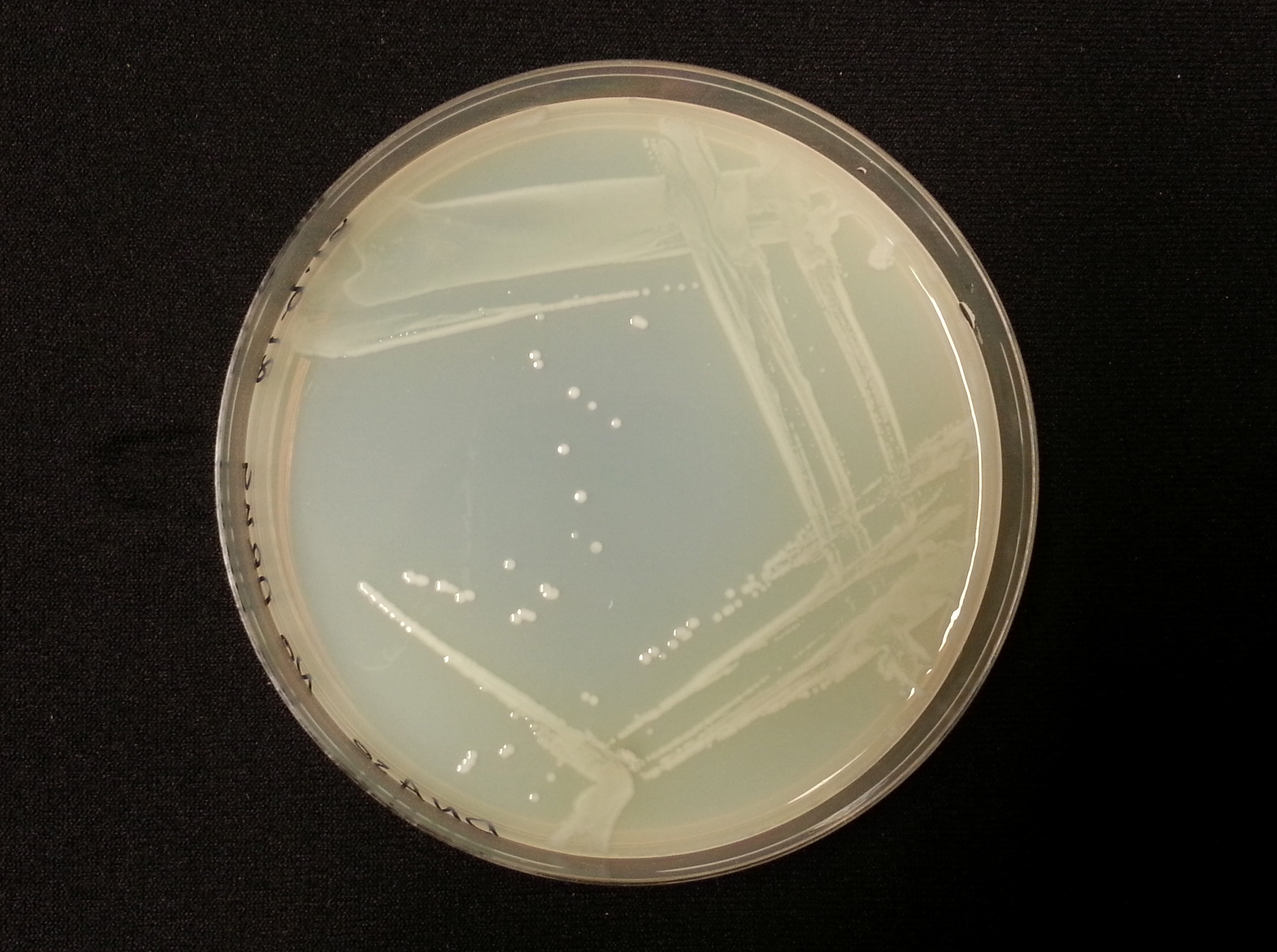 | 
|
 "
"