Team:ZJU-China/models.htm
From 2012.igem.org
(Difference between revisions)
| Line 288: | Line 288: | ||
<p align="justify">In this year’s project, we try to make reasonable design of riboscaffolds by ourselves. Due to the current experimental conditions, we could not know what exactly happens to our riboscaffolds in the cell. Its folding process, binding with proteins and ligand, and the allosteric transition are of interests, since these are considerations for a reasonable design. Luckily, we have simulation tools for molecular modeling which can provide hints about what may happen theoretically in the intracellular environment. Although molecular modeling is often considered as professional work in which knowledge of different disciplines such as biology, physics, chemistry, mathematics and computer science as well as previous experience all contribute to good results, our team is willing to take a step in this field and get preliminary results.</p> | <p align="justify">In this year’s project, we try to make reasonable design of riboscaffolds by ourselves. Due to the current experimental conditions, we could not know what exactly happens to our riboscaffolds in the cell. Its folding process, binding with proteins and ligand, and the allosteric transition are of interests, since these are considerations for a reasonable design. Luckily, we have simulation tools for molecular modeling which can provide hints about what may happen theoretically in the intracellular environment. Although molecular modeling is often considered as professional work in which knowledge of different disciplines such as biology, physics, chemistry, mathematics and computer science as well as previous experience all contribute to good results, our team is willing to take a step in this field and get preliminary results.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>RNA folding and 3D Structures</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<h5>Introduction</h5> | <h5>Introduction</h5> | ||
| Line 338: | Line 338: | ||
<p align="justify">To demonstrate that the interaction in Clover3 is caused by our design, we simulate the 3D structure of a mutant of Clover 3, in which the theophylline aptamer remains unmutant so it cannot base pair with the sequence of MS2.</p> | <p align="justify">To demonstrate that the interaction in Clover3 is caused by our design, we simulate the 3D structure of a mutant of Clover 3, in which the theophylline aptamer remains unmutant so it cannot base pair with the sequence of MS2.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>Docking</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<h5>Introduction</h5> | <h5>Introduction</h5> | ||
| Line 372: | Line 372: | ||
<p align="justify">The docking results were used to generate riboscaffold-theophylline complex for molecular dynamics analysis of the whole system.</p> | <p align="justify">The docking results were used to generate riboscaffold-theophylline complex for molecular dynamics analysis of the whole system.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>Molecular Dynamics</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<h5>Introduction</h5> | <h5>Introduction</h5> | ||
| Line 444: | Line 444: | ||
<p align="justify">In conclusion, the preliminary analysis results show that the simulation system generated by our workflow acts similar to other ligand-receptor systems under the environment of molecular dynamics. Actually, with this method, we can conduct more analysis on the level of molecules and even atoms. We will continue our way to explore this magic microworld!</p> | <p align="justify">In conclusion, the preliminary analysis results show that the simulation system generated by our workflow acts similar to other ligand-receptor systems under the environment of molecular dynamics. Actually, with this method, we can conduct more analysis on the level of molecules and even atoms. We will continue our way to explore this magic microworld!</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>References</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">[1] Yaroslav Chushak, Morley O. Stone. In silico selection of RNA aptamers. Nucl. Acids Res. (2009) 37(12).( http://nar.oxfordjournals.org/content/37/12/e87.short)</p> | <p align="justify">[1] Yaroslav Chushak, Morley O. Stone. In silico selection of RNA aptamers. Nucl. Acids Res. (2009) 37(12).( http://nar.oxfordjournals.org/content/37/12/e87.short)</p> | ||
| Line 479: | Line 479: | ||
<div class="acc_container" style="display: none; "> | <div class="acc_container" style="display: none; "> | ||
<!--Content Goes Here--> | <!--Content Goes Here--> | ||
| - | < | + | <h2>Introduction</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">As explained before, the RNA scaffold brings the two enzymes (MS2 and PP7) closer, which improve the reaction rate of pathways. It seems magical that the reaction rate would be improved when two enzymes are close to each other. Then some questions are emerging. How faster it would be when two enzymes are in a RNA scaffold? What are the factors that will affect the efficiency of pathways? Those are what we want to discuss below. In this section, we establish a model by simplifying the molecular motion and simulating the reaction using Matlab programming.</p> | <p align="justify">As explained before, the RNA scaffold brings the two enzymes (MS2 and PP7) closer, which improve the reaction rate of pathways. It seems magical that the reaction rate would be improved when two enzymes are close to each other. Then some questions are emerging. How faster it would be when two enzymes are in a RNA scaffold? What are the factors that will affect the efficiency of pathways? Those are what we want to discuss below. In this section, we establish a model by simplifying the molecular motion and simulating the reaction using Matlab programming.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>Simplification</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">We simplify our abstract model as a pathway A -> B -> C. Enzyme one (E1) catalyzes the reaction A -> B and enzyme two (E2) catalyzes the reaction B -> C. The reaction equation is:</p> | <p align="justify">We simplify our abstract model as a pathway A -> B -> C. Enzyme one (E1) catalyzes the reaction A -> B and enzyme two (E2) catalyzes the reaction B -> C. The reaction equation is:</p> | ||
| Line 491: | Line 491: | ||
<p align="justify">Molecular motion is continuous and complex so that it is difficult to model precisely. As a simplification, it is reasonable to discretize the continuous time as discrete steps and simplify the molecular Brownian motion as three-dimensional random walking. With this simplification, we can approximately observe the process of reaction and the reaction rate. Therefore, we present algorithms to simulate the process of molecular reaction dynamically and intuitionally.</p> | <p align="justify">Molecular motion is continuous and complex so that it is difficult to model precisely. As a simplification, it is reasonable to discretize the continuous time as discrete steps and simplify the molecular Brownian motion as three-dimensional random walking. With this simplification, we can approximately observe the process of reaction and the reaction rate. Therefore, we present algorithms to simulate the process of molecular reaction dynamically and intuitionally.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>Algorithm</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">In order to simulate the molecular motion and verify the fact that a RNA scaffold with two enzymes can speed reactions of the pathway, we simplify the complex realistic situation as a simple and virtual three dimensional world.</p> | <p align="justify">In order to simulate the molecular motion and verify the fact that a RNA scaffold with two enzymes can speed reactions of the pathway, we simplify the complex realistic situation as a simple and virtual three dimensional world.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>Assumptions:</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">1. The container is a cube and we assumed a unit. The edge of the cube is an integral multiple of one unit.</p> | <p align="justify">1. The container is a cube and we assumed a unit. The edge of the cube is an integral multiple of one unit.</p> | ||
| Line 505: | Line 505: | ||
<p align="justify">4. The molecule cannot cross the edge of the cube.</p> | <p align="justify">4. The molecule cannot cross the edge of the cube.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>Algorithm:</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">Initial state: All the molecules including A, E1 and E2 are randomly scattered at the intersections</p> | <p align="justify">Initial state: All the molecules including A, E1 and E2 are randomly scattered at the intersections</p> | ||
| Line 515: | Line 515: | ||
<p align="justify">Step 3: All the molecules are randomly walking one unit. If the molecule is going to across the border, it will be still and does not have a walk. Then, go back to step 1.</p> | <p align="justify">Step 3: All the molecules are randomly walking one unit. If the molecule is going to across the border, it will be still and does not have a walk. Then, go back to step 1.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>Implementation</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">We use the Matlab programming to construct the model and implement this algorithm, presenting the results using both data and 3D figures.</p> | <p align="justify">We use the Matlab programming to construct the model and implement this algorithm, presenting the results using both data and 3D figures.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>A small scale simulation</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
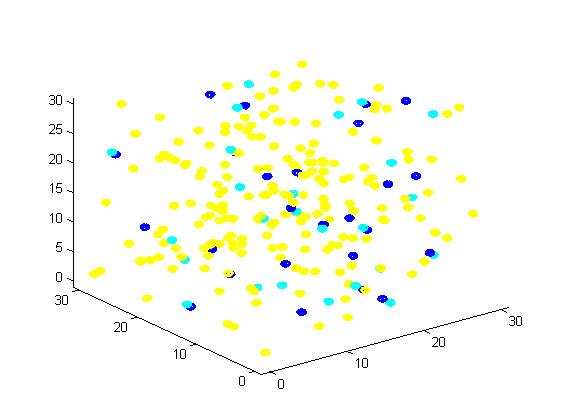
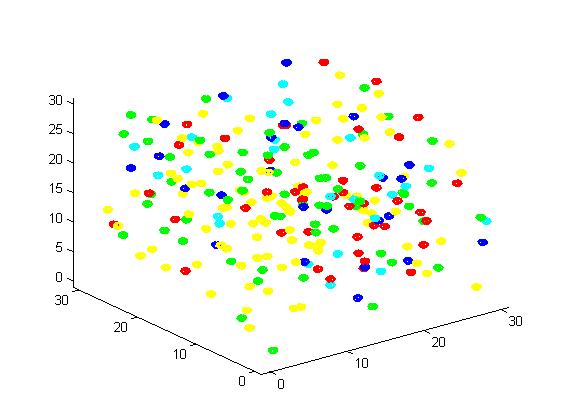
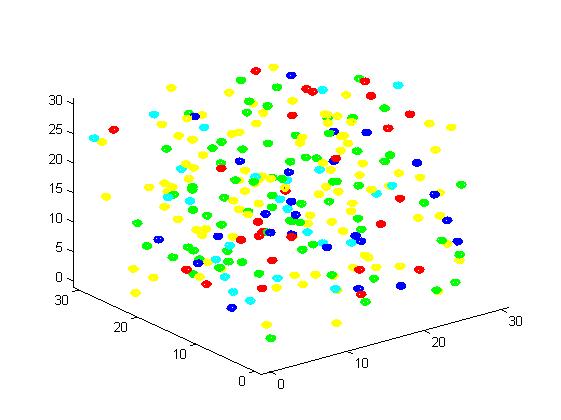
<p align="justify">A small scale simulation is used to verify our algorithm, providing a clearly recognize of our algorithm. </p> | <p align="justify">A small scale simulation is used to verify our algorithm, providing a clearly recognize of our algorithm. </p> | ||
| Line 552: | Line 552: | ||
<p align="justify">From the results, we can find that the number of C for scaffold is more than those for non-scaffold. Therefore, enzyme one and enzyme two being on a RNA scaffold can help to improve the reaction rate. The figures can help to provide a visible reflection. Though the small scale simulation has verified our algorithm, we also present a large scale simulation and have some discussion.</p> | <p align="justify">From the results, we can find that the number of C for scaffold is more than those for non-scaffold. Therefore, enzyme one and enzyme two being on a RNA scaffold can help to improve the reaction rate. The figures can help to provide a visible reflection. Though the small scale simulation has verified our algorithm, we also present a large scale simulation and have some discussion.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>A large scale simulation</h2> |
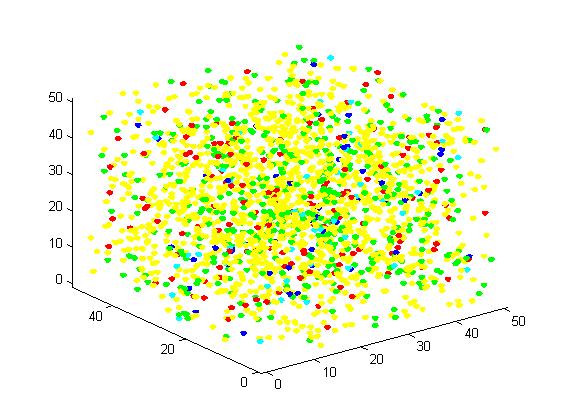
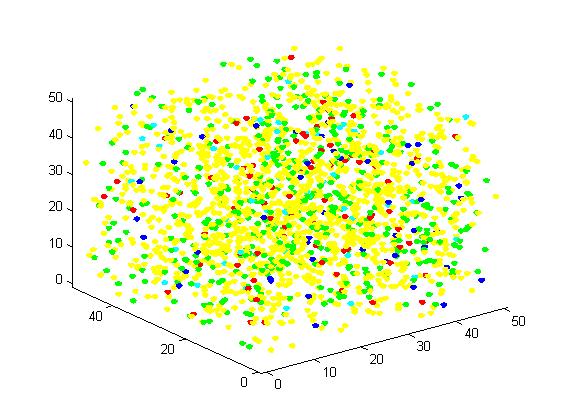
<p align="justify">The initial state is as following:</p> | <p align="justify">The initial state is as following:</p> | ||
<p align="justify">Edge of cube: 50 units</p> | <p align="justify">Edge of cube: 50 units</p> | ||
| Line 574: | Line 574: | ||
<p align="justify">Figure 5. Result of non-scaffold system for large scale simulation. After 1200 iterations, there are 451 B and 79 C.</p> | <p align="justify">Figure 5. Result of non-scaffold system for large scale simulation. After 1200 iterations, there are 451 B and 79 C.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>Comparison between scaffold system and non-scaffold system</h2> |
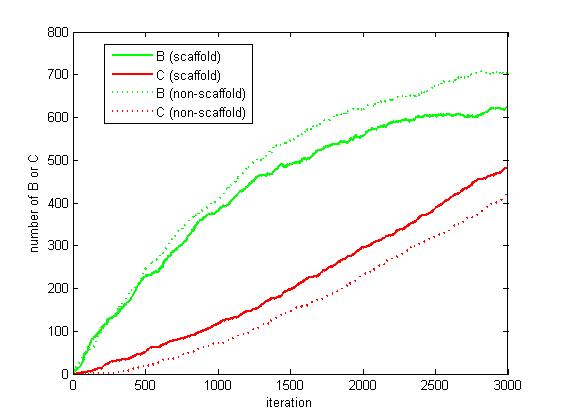
<p align="justify">As the iteration increasing, the number of A is decreasing and the number of B and C are increasing. We record the number of B and C for both scaffold system and non-scaffold system as iteration grows to watch the reaction rate of pathway (i.e. the number of C).</p> | <p align="justify">As the iteration increasing, the number of A is decreasing and the number of B and C are increasing. We record the number of B and C for both scaffold system and non-scaffold system as iteration grows to watch the reaction rate of pathway (i.e. the number of C).</p> | ||
<div><img src="http://www.jiajunlu.com/igem/zju_model2_6.jpg"></div> | <div><img src="http://www.jiajunlu.com/igem/zju_model2_6.jpg"></div> | ||
| Line 581: | Line 581: | ||
<p align="justify">From figure 4.5.6, it is obvious that the number of C for scaffold system is more than that for non-scaffold system and the number of B for scaffold system is less than that for non-scaffold system. This phenomenon can be interpreted as that the probability that B meets an E2 is highly increased since their distance is closer. Therefore, it is safely to draw a conclusion that the reaction rate of pathway has been speed up because of the scaffold bringing two enzymes closer.</p> | <p align="justify">From figure 4.5.6, it is obvious that the number of C for scaffold system is more than that for non-scaffold system and the number of B for scaffold system is less than that for non-scaffold system. This phenomenon can be interpreted as that the probability that B meets an E2 is highly increased since their distance is closer. Therefore, it is safely to draw a conclusion that the reaction rate of pathway has been speed up because of the scaffold bringing two enzymes closer.</p> | ||
| - | < | + | <h2>Effect of enzymes concentration</h2> |
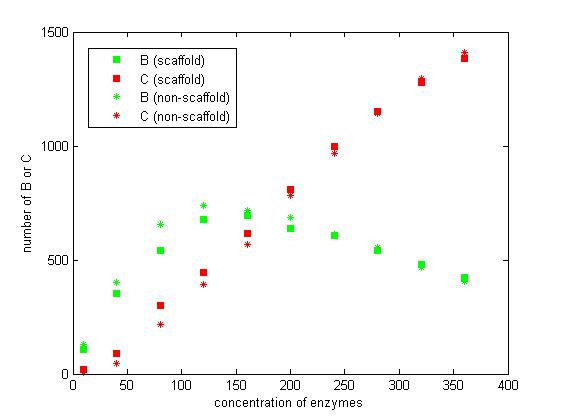
<p align="justify">From the above discussion, we assumed the concentration of enzymes is constant. However, the increasing concentration of enzymes also results in the increasing of reaction rate. The difference between scaffold and non-scaffold system varies when the concentration of enzymes increase.</p> | <p align="justify">From the above discussion, we assumed the concentration of enzymes is constant. However, the increasing concentration of enzymes also results in the increasing of reaction rate. The difference between scaffold and non-scaffold system varies when the concentration of enzymes increase.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| Line 590: | Line 590: | ||
<p align="justify">From the figure 7, we find that when the concentration of enzymes is too low and too high, the difference between scaffold and non-scaffold system is not noticeable. Only when the concentration of enzymes is at a certain range of value, the advantages of scaffold system is significant. It seems reasonable that when the concentration is too low, the reaction rate is also low. When the concentration is too high, the probability that B meets an E2 is approximately equal for both systems.</p> | <p align="justify">From the figure 7, we find that when the concentration of enzymes is too low and too high, the difference between scaffold and non-scaffold system is not noticeable. Only when the concentration of enzymes is at a certain range of value, the advantages of scaffold system is significant. It seems reasonable that when the concentration is too low, the reaction rate is also low. When the concentration is too high, the probability that B meets an E2 is approximately equal for both systems.</p> | ||
| - | < | + | <h2>Effect of distance between two enzymes</h2> |
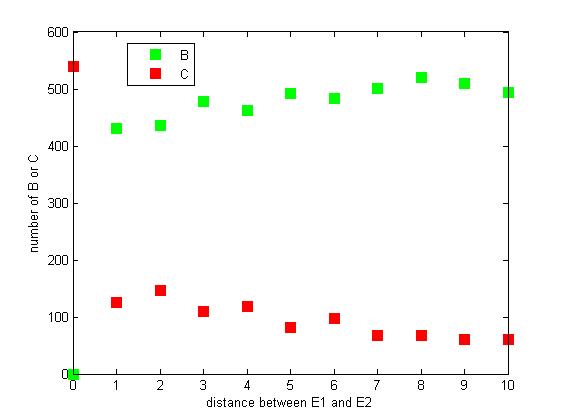
<p align="justify">The distance between two enzymes E1 and E2 on RNA scaffold has an influence on reaction rate of pathway. If the E1 and E2 are closer, the probability that B, that is generated from A, meets an E2 is high for the reason that the distance between B and E2 are close. When the distance between E1 and E2 are getting further, the number of C is declining.</p> | <p align="justify">The distance between two enzymes E1 and E2 on RNA scaffold has an influence on reaction rate of pathway. If the E1 and E2 are closer, the probability that B, that is generated from A, meets an E2 is high for the reason that the distance between B and E2 are close. When the distance between E1 and E2 are getting further, the number of C is declining.</p> | ||
<div><img src="http://www.jiajunlu.com/igem/zju_model2_8.jpg"></div> | <div><img src="http://www.jiajunlu.com/igem/zju_model2_8.jpg"></div> | ||
| Line 603: | Line 603: | ||
<div class="acc_container" style="display: none; "> | <div class="acc_container" style="display: none; "> | ||
<!--Content Goes Here--> | <!--Content Goes Here--> | ||
| - | < | + | <h2>Introduction </h2> |
<p align="justify">The binding of a RNA aptamer to MS2 or PP7 is dynamic, which means some MS2 and PP7 are binding with RNA aptamers and others are separated from RNA aptamers. Having known the initial concentration of RNA aptamer, MS2 and PP7, we need to find out how many RNA scaffolds have both MS2 and PP7 when the binding reach a final equilibrium. In this section, we use the concept -- association rate and dissociation rate [1] to model the process of binding.</p> | <p align="justify">The binding of a RNA aptamer to MS2 or PP7 is dynamic, which means some MS2 and PP7 are binding with RNA aptamers and others are separated from RNA aptamers. Having known the initial concentration of RNA aptamer, MS2 and PP7, we need to find out how many RNA scaffolds have both MS2 and PP7 when the binding reach a final equilibrium. In this section, we use the concept -- association rate and dissociation rate [1] to model the process of binding.</p> | ||
| - | < | + | <h2>Assumption</h2> |
<p align="justify">There are two aptamers on the RNA scaffold named aptamer1 (A1) and aptamer2 (A2). Aptamer1 is binding with MS2 and aptamer2 is binding with PP7. Before establishing the model, we declare some assumptions.</p> | <p align="justify">There are two aptamers on the RNA scaffold named aptamer1 (A1) and aptamer2 (A2). Aptamer1 is binding with MS2 and aptamer2 is binding with PP7. Before establishing the model, we declare some assumptions.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| Line 633: | Line 633: | ||
<p align="justify">[`K_{-2}`] : Dissociation rate of aptamer2 to PP7</p> | <p align="justify">[`K_{-2}`] : Dissociation rate of aptamer2 to PP7</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>Differential equation modeling</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">The binding of aptamer1 to MS2 and aptamer2 to PP7 can be presented as reaction formula:</p> | <p align="justify">The binding of aptamer1 to MS2 and aptamer2 to PP7 can be presented as reaction formula:</p> | ||
| Line 660: | Line 660: | ||
<p align="justify">`[PP7A2_0]=0`</p> | <p align="justify">`[PP7A2_0]=0`</p> | ||
| - | < | + | <h2>Equilibrium analysis</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
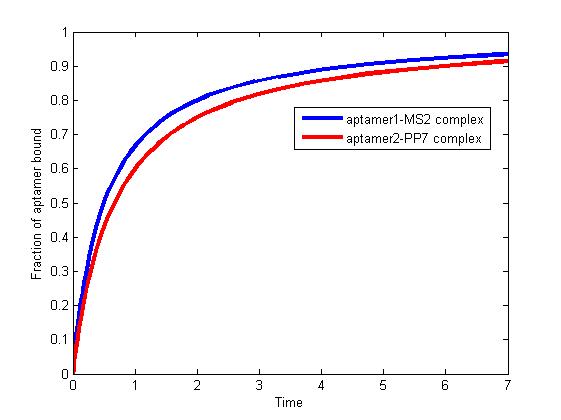
<p align="justify">We solve the differential equations numerically using the "ode" solver in Matlab. Titolo et at once come up with an approach to determine the equilibrium dissociation constant. This constant is dependent on anisotropies of the sample, aptamer, and intensities of the polarizers[1]. Therefore, the association rate constant and dissociation rate constant are usually experimentally determined values. It is difficult for us to determine the constants precisely since they vary in terms of experimental environment. </p>According to literature, we assumed that</p> | <p align="justify">We solve the differential equations numerically using the "ode" solver in Matlab. Titolo et at once come up with an approach to determine the equilibrium dissociation constant. This constant is dependent on anisotropies of the sample, aptamer, and intensities of the polarizers[1]. Therefore, the association rate constant and dissociation rate constant are usually experimentally determined values. It is difficult for us to determine the constants precisely since they vary in terms of experimental environment. </p>According to literature, we assumed that</p> | ||
| Line 675: | Line 675: | ||
<p align="justify">As we mentioned above, the binding of MS2 to aptamer1 and the binding of PP7 to aptamer2 are independent with each other. The binding of one would not affect the binding of the other. From probability theoretical perspective, the fraction of RNA scaffold binding with both MS2 and PP7 is equal to the fraction of RNA scaffold binding with MS2 multiply the fraction of RNA scaffold binding with PP7. The RNA scaffold binding with both MS2 and PP7 is what we need to improve the reaction rate of pathways. </p> | <p align="justify">As we mentioned above, the binding of MS2 to aptamer1 and the binding of PP7 to aptamer2 are independent with each other. The binding of one would not affect the binding of the other. From probability theoretical perspective, the fraction of RNA scaffold binding with both MS2 and PP7 is equal to the fraction of RNA scaffold binding with MS2 multiply the fraction of RNA scaffold binding with PP7. The RNA scaffold binding with both MS2 and PP7 is what we need to improve the reaction rate of pathways. </p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| - | < | + | <h2>References</h2> |
<p align="justify">[1] Ajish S. R. Potty, Katerina Kourentzi, Han Fang, George W. Jackson, Xing Zhang, Glen B. Legge, Richard C. Willson. Biophysical Characterization of DNA Aptamer Interactions with Vascular Endothelial Growth Factor. 2008.</p> | <p align="justify">[1] Ajish S. R. Potty, Katerina Kourentzi, Han Fang, George W. Jackson, Xing Zhang, Glen B. Legge, Richard C. Willson. Biophysical Characterization of DNA Aptamer Interactions with Vascular Endothelial Growth Factor. 2008.</p> | ||
</div><!-- end .acc_container --> | </div><!-- end .acc_container --> | ||
Revision as of 09:29, 26 September 2012

 "
"