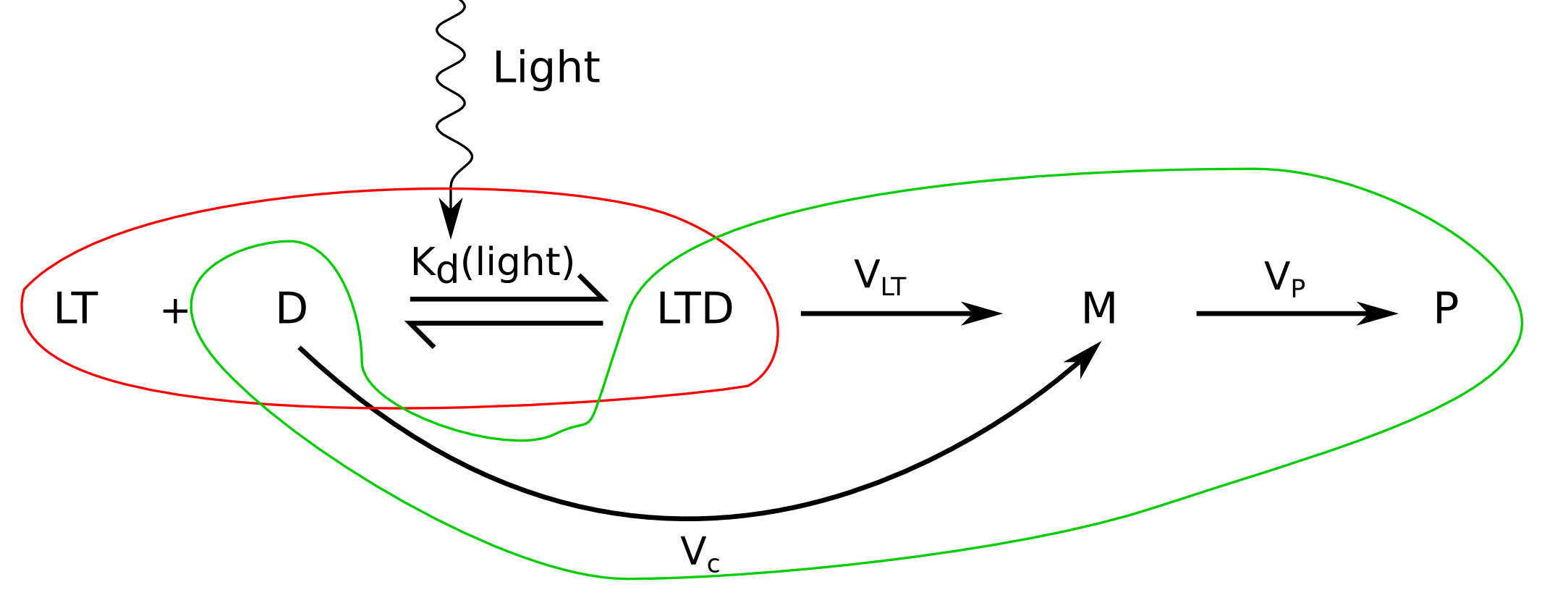
Why?
From our work on LOV2 photoactivation we might be able to predict the percentage of LOV2 domains in our LovTAP-VP16, even though this value might be affected by the fact of having both ends fused to other domains: TrpR and VP16. However, some authors, as Kasahara et al, worked with fusion LOV2 proteins, getting comparable parameters.
We will then assume that we know the proportion of activated LovTAP-VP16 monomers. But LovTAP-VP16 has to dimerize to be able to bind DNA. In the section [[Team:EPF-Lausanne/Modeling/
92% of photoactive LOV2 has it's Jα-helix undocked (Yao et al, 2008).
Eitoku et al (2005) say it takes 2 ms for the J&alpha-helix to undock.
 "
"