Team:SJTU-BioX-Shanghai/Project/project1.2
From 2012.igem.org
AleAlejandro (Talk | contribs) (→Construction) |
AleAlejandro (Talk | contribs) (→Construction) |
||
| Line 41: | Line 41: | ||
|_''Fig.1 :'' PDZ domain & ligand __|_''Fig.2 :'' GBD domain & ligand __|___''Fig.3 :'' SH3 domain & ligand__| | |_''Fig.1 :'' PDZ domain & ligand __|_''Fig.2 :'' GBD domain & ligand __|___''Fig.3 :'' SH3 domain & ligand__| | ||
| - | |____ from PDB 2PDZ ___________|_______ from PDB | + | |____ from PDB ''2PDZ'' ___________|_______ from PDB ''1T84''_________|________ from PDB ''1CKB''_________| |
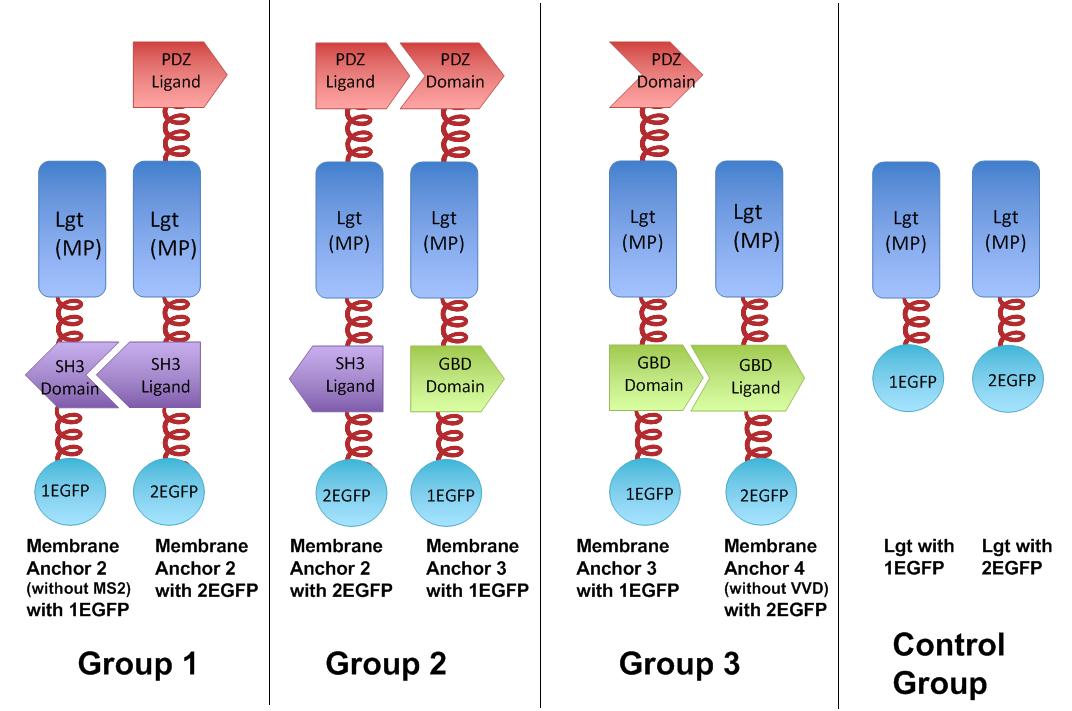
| - | As demonstrated before, we built our device on E.coli inner membrane protein Lgt. SRP-dependent signaling sequence of DsbA is fused to N-terminus to localize fusion proteins to membrane. | + | As demonstrated before, we built our device on E.coli inner membrane protein Lgt. SRP-dependent signaling sequence of DsbA is fused to N-terminus to localize fusion proteins to membrane. |
| - | To assemble and arrange enzymes onto inner membrane of E.coli, we incorporate three protein domains that can interact with their cognate ligands. (Dueber, Wu et al. 2009) Protein-protein interaction domains and ligands from metazoan cells (mouse SH3 and PDZ domains and rat GBD domain) were recruited to assemble enzymes on membrane. Each domain could assemble with its cognate ligand. | + | To assemble and arrange enzymes onto inner membrane of E.coli, we incorporate three protein domains that can interact with their cognate ligands. (Dueber, Wu et al. 2009) Protein-protein interaction domains and ligands from metazoan cells (mouse SH3 and PDZ domains and rat GBD domain) were recruited to assemble enzymes on membrane. Each domain could assemble with its cognate ligand. |
===Fluorescence Complementation Assay=== | ===Fluorescence Complementation Assay=== | ||
Revision as of 07:27, 26 September 2012
| ||
|
 "
"