Team:LMU-Munich/Bacillus BioBricks/vector use
From 2012.igem.org
| Line 36: | Line 36: | ||
To check if the plasmid was taken up, the transformed ''B. subtilis'' is plated on selective media, LB with the appropriate antibiotic (resistance gene in between recombination sites). The obtained colonies then are tested for their insertion into the correct locus. Usually it is sufficient to test 4-8 colonies. | To check if the plasmid was taken up, the transformed ''B. subtilis'' is plated on selective media, LB with the appropriate antibiotic (resistance gene in between recombination sites). The obtained colonies then are tested for their insertion into the correct locus. Usually it is sufficient to test 4-8 colonies. | ||
</p> | </p> | ||
| - | + | ||
To test the insertion into the | To test the insertion into the | ||
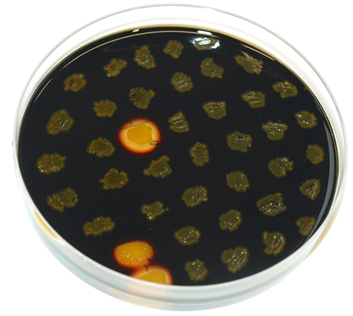
| - | * amyE-locus: streak the obtained colonies (and the WT as control) on a replica plate (with antibiotic) and on a starch plate and let grow overnight at 37°C. The next day, pour Lugol’s iodine on the plate so that it is covered with a thin film. Around colonies which can degrade starch (WT and wrong colonies), there should be a bright zone around the colony. Correct clones do not show this bright zone. (see also Figure 1) | + | * <p align="justify">amyE-locus: streak the obtained colonies (and the WT as control) on a replica plate (with antibiotic) and on a starch plate and let grow overnight at 37°C. The next day, pour Lugol’s iodine on the plate so that it is covered with a thin film. Around colonies which can degrade starch (WT and wrong colonies), there should be a bright zone around the colony. Correct clones do not show this bright zone. (see also Figure 1) </p> |
{| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| Line 53: | Line 53: | ||
|} | |} | ||
| - | + | ||
| - | <p align="justify"> | + | |
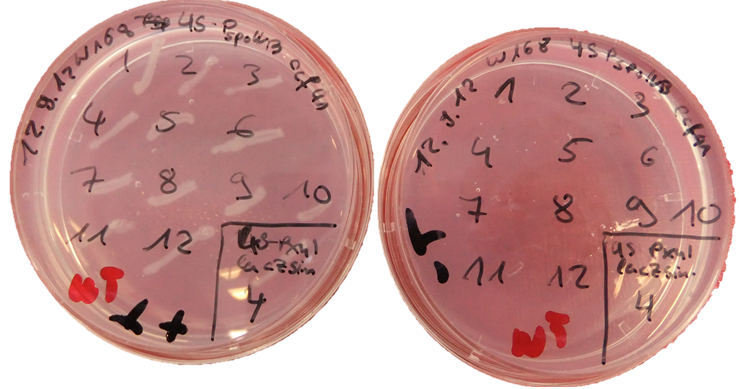
| - | + | * <p align="justify">thrC-locus: streak the obtained colonies (and the WT as control) on a replica plate (with antibiotic) and on a plate with minimal medium without threonine and minimal medium with threonine (use the MNGE media, recipe see transformation protocol). Correct colonies should grow only on the LB and minimal medium with threonine plate (see Figure 2).</p> | |
| - | </p> | + | |
{| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| style="width: 70%;background-color: #EBFCE4;" | | | style="width: 70%;background-color: #EBFCE4;" | | ||
| Line 73: | Line 73: | ||
|} | |} | ||
| - | <p align="justify"> | + | |
| - | + | * <p align="justify">sacA-locus and lacA-locus: for those two loci, a colony PCR should be performed. The protocol can be found on our website.</p> | |
| - | * For sacA: you can use the primers: up: CTGATTGGCATGGCGATTGC together with ACAGCTCCAGATCCTCTACG as well as down: GTCGCTACCATTACCAGTTG together with TCCAAACATTCCGGTGTTATC. | + | * <p align="justify">For sacA: you can use the primers: up: CTGATTGGCATGGCGATTGC together with ACAGCTCCAGATCCTCTACG as well as down: GTCGCTACCATTACCAGTTG together with TCCAAACATTCCGGTGTTATC. |
</p> | </p> | ||
| - | <p align="justify"> | + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" |
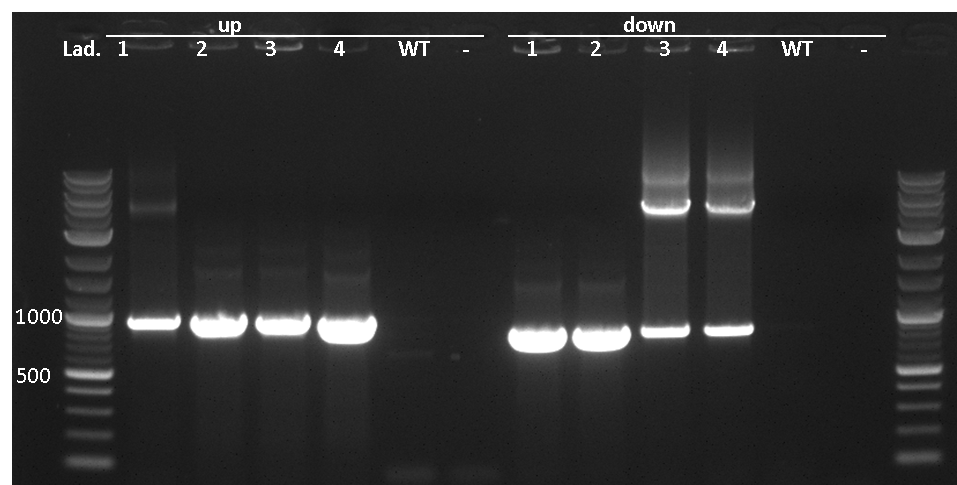
| - | Figure 3: Colony PCR of pSBBs3C-luxABCDE-PlepA. The expected bands are: up: 946 bp, down: 930 bp, none in WT and none with water as negative control. So all of the checked colonies have the insertion in the right locus. | + | | style="width: 70%;background-color: #EBFCE4;" | |
| - | * for lacA, you can use the primers: [not designed yet] | + | {| |
| + | |[[File:LMU-Munich-Gelfoto colony PCR.png|500px|center]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2"><p align="justify"> | ||
| + | Figure 3: Colony PCR of pSBBs3C-luxABCDE-PlepA. The expected bands are: up: 946 bp, down: 930 bp, none in WT and none with water as negative control. So all of the checked colonies have the insertion in the right locus.</p></font> | ||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | |||
| + | * <p align="justify"> for lacA, you can use the primers: [not designed yet]</p> | ||
With colony PCR you can check any other locus. | With colony PCR you can check any other locus. | ||
| - | + | ||
Revision as of 11:16, 25 September 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

How to work with Bacillus subtilis vectors
There are some features in B. subtilis vectors that have to be taken into account, while working with them.
- Cloning in E.coli
- Linearisation before B. subtilis transformation
- Verification of integration
Pre-Cloning in E. coli
The cloning, that means the insertion of your part into the multiple cloning site, is done with normal ligations and E. coli. However, since the B. subtilis vectors are quite large, the cloning works best if only one insert is inserted. You could try to finish your construct in e.g. pSB1C3 and then clone it into the B. subtilis vector. For convenience, all vectors carry an RFP with promoter and terminator which is substituted by your insert during the ligation.
Our vectors all carry the bla gene that mediates Ampicillin resistance (100µg/ml) in E. coli. Also, they all have two recombination sites to result in a double crossover in a designated locus. In between those recombination sites, there is the multiple cloning site and a resistance marker for B. subtilis.
(Some vectors from other working groups do not carry an extra E. coli resistance, so the B. subtilis resistance is also used in E. coli but with lower antibiotic concentrations. There also is the possibility of single-crossover plasmids which do also work fine, but the vector can then easily cross-out again, so it is not stably integrated.)
Linearisation before transformation in B. subtilis
Nevertheless, the obtained plasmid (if it is not replicative) has to be linearized before transformation. B. subtilis is naturally competent, but preferably takes up and integrates linear DNA fragments. The linearization of our plasmids can all be performed with ScaI which only cuts inside the bla gene. If that enzyme also cuts in your insert, please check for other single cutters outside of the area that is integrated into the B. subtilis genome. For the actual transformation of B. subtilis, please linearize 1-2 µg of your plasmid and then proceed with our transformation protocol.
Verification of correct integration
To check if the plasmid was taken up, the transformed B. subtilis is plated on selective media, LB with the appropriate antibiotic (resistance gene in between recombination sites). The obtained colonies then are tested for their insertion into the correct locus. Usually it is sufficient to test 4-8 colonies.
To test the insertion into the
-
amyE-locus: streak the obtained colonies (and the WT as control) on a replica plate (with antibiotic) and on a starch plate and let grow overnight at 37°C. The next day, pour Lugol’s iodine on the plate so that it is covered with a thin film. Around colonies which can degrade starch (WT and wrong colonies), there should be a bright zone around the colony. Correct clones do not show this bright zone. (see also Figure 1)
|
-
thrC-locus: streak the obtained colonies (and the WT as control) on a replica plate (with antibiotic) and on a plate with minimal medium without threonine and minimal medium with threonine (use the MNGE media, recipe see transformation protocol). Correct colonies should grow only on the LB and minimal medium with threonine plate (see Figure 2).
|
-
sacA-locus and lacA-locus: for those two loci, a colony PCR should be performed. The protocol can be found on our website.
-
For sacA: you can use the primers: up: CTGATTGGCATGGCGATTGC together with ACAGCTCCAGATCCTCTACG as well as down: GTCGCTACCATTACCAGTTG together with TCCAAACATTCCGGTGTTATC.
|
-
for lacA, you can use the primers: [not designed yet]
With colony PCR you can check any other locus.
 "
"