Team:Technion/Project/Overview
From 2012.igem.org
(→The general idea) |
(→The general idea) |
||
| Line 1: | Line 1: | ||
{{:Team:Technion/Project}} | {{:Team:Technion/Project}} | ||
==The general idea== | ==The general idea== | ||
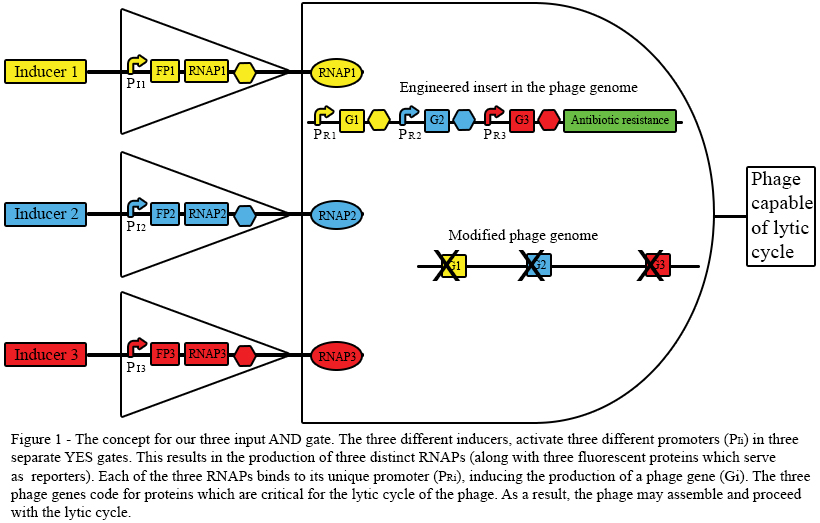
| - | Figure 1 presents the basic strategy for our three inputs AND gate. Each one of the inducers can be an environmental signal, to which the phage will respond. In order to check that the inducers work, we chose to use fluorescent proteins (FPs). The inducers also trigger the production of RNA polymerases (RNAPs), in a logical operation similar to a [https://2012.igem.org/Team:Technion/Project/YES_gates#What.27s_a_YES_gate.3F YES gate]. Finally, in the presence of all the inputs, the phage will be capable to resume its lytic cycle, resulting in our AND gate's output.<br> | + | Figure 1 presents the basic strategy for our three inputs AND gate. Each one of the inducers can be an environmental signal, to which the phage will respond. In order to check that the inducers work, we chose to use fluorescent proteins (FPs) as reporters. The inducers also trigger the production of RNA polymerases (RNAPs), in a logical operation similar to a [https://2012.igem.org/Team:Technion/Project/YES_gates#What.27s_a_YES_gate.3F YES gate]. Finally, in the presence of all the inputs, the phage will be capable to resume its lytic cycle, resulting in our AND gate's output.<br> |
[[File:Technion_overview.jpg|800px|thumb|center]] | [[File:Technion_overview.jpg|800px|thumb|center]] | ||
Revision as of 15:40, 24 September 2012

Contents |
The general idea
Figure 1 presents the basic strategy for our three inputs AND gate. Each one of the inducers can be an environmental signal, to which the phage will respond. In order to check that the inducers work, we chose to use fluorescent proteins (FPs) as reporters. The inducers also trigger the production of RNA polymerases (RNAPs), in a logical operation similar to a YES gate. Finally, in the presence of all the inputs, the phage will be capable to resume its lytic cycle, resulting in our AND gate's output.
The inducers, promoters and bacterial strain
The inducers serve as the inputs for the three YES gates. When considering the choice of our system's components, the choice of the inducers, promoters and bacterial strain is intertwined. We needed to minimize the number of plasmids used, therefore, the bacterial strain should provide us with as many regulatory overexpressed components as possible. Roee had the Escherichia coli (E. coli) strain DH5αZ1, which expresses high levels of Lac and Tet repressor (LacR, TetR) as well as of AraC {1}. This allowed us to use the PTetO and the PLac/Ara promoters without the introduction of additional regulatory elements on plasmids. Consequentially, the inducers used were anhydrotetracycline (aTc) for PTetO and IPTG with arabinose for PLac/Ara.
The choice of the third promoter could no longer rely on host strain expression of required regulatory elements. Therefore, we chose the quorum sensing system based on the PLux promoter, which is induced by [http://partsregistry.org/3OC6HSL 3OC6HSL] (also known as N-(β-ketocaproyl)-L-Homoserine lactone) in the presence of the LuxR protein. This made it obligatory for us to express the LuxR gene from a plasmid. Moreover, since the PLux promoter is leaky, we chose to introduce another regulatory element to reduce the basal level. This element is the theophylline induced riboswitch (RS), which controls the level of translation of a given protein. This construct is later notated as PLux+RS. More about these elements can be found in the YES gate #3 section.
The fluorescent proteins
The purpose of the fluorescent proteins in our system was to verify induction of the various promoters by their inducers. The choice of compatible fluorescent proteins for simultaneous visualization can be tricky. Based on the work done by Tsien, et al {2} we initially chose to use mCitrine (a yellow FP), Cerulean (a cyan FP) and mCherry (a red FP) assigned to PTetO, PLac/Ara and PLux+RS respectively. Following different consideration which are mentioned in the YES gates section for each system. Unfortunately, the mCitrine, which we acquired from Roee, failed to show fluorescence. Therefore, we tried several alternatives which are described in the YES gate #1 section.
The RNA polymerases and promoters
The output from each of our YES gates is the production of a distinct RNA polymerase. When choosing the polymerases for our system, a few key features were kept in mind:
- Orthogonality - each of the RNAPs used should be orthogonal to the other components of the circuit. Crosstalk between the different RNAPs should be minimal. Thus, reducing the chance of a false positive result.
- Host compatibility - the chosen RNAPs should be compatible with our chosen host strain on several accounts. Firstly, there should be no crosstalk between the host RNAPs and the chosen promoters. Moreover, the chosen RNAPs should not burden the cell or be toxic to it.
- Tuneable strength - the chosen RNAPs trigger expression from dedicated promoters. It should be possible to tune the level of expression by choosing a specific promoter from a promoter library corresponding to each RNAP
In order to answer the above demands, bacteriophage RNAPs and promoter libraries were chosen. More information about the different RNAPs can be found in the RNAPs section.
The phage
The final phase in our system's logic operations, is the AND gate operation itself. The AND function is achieved using an engineered phage. The chosen strain for our project is the heat inducible cI857s7 lambda phage, which attacks E. coli cells. To achieve the AND logic operation, the phage must be engineered by gene deletions and insertions under our RNAP promoters. More information about the phage strain and the genetic modifications to it is available in the phage section.
References
1. Lutz R., Bujard H. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Research 25(6): 1203-1210.
2. Shaner N. C., Steinbach P. A., Tsien R. Y. 2005. A guide to choosing fluorescent proteins. Nature Methods 2(12): 905-909.
 "
"