Team:Kyoto/Florigen/Project
From 2012.igem.org
(→Construction) |
(→Construction) |
||
| Line 19: | Line 19: | ||
==1. Expression of FT in E.coli== | ==1. Expression of FT in E.coli== | ||
==Construction== | ==Construction== | ||
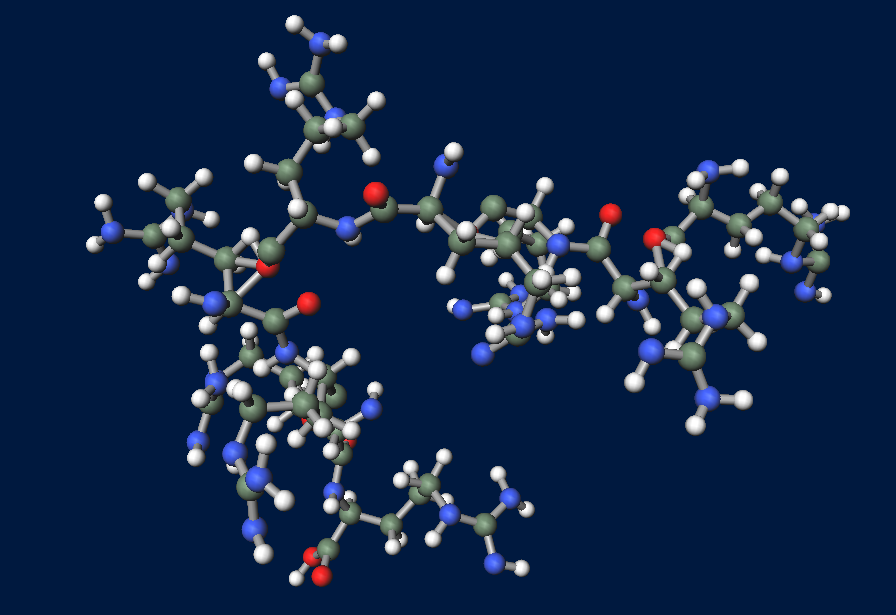
| - | FT gene has 2 BioBrick restriction enzyme sites, EcoR1 and Pst1(fig.3A.) So we did mutation to delete the sites before we start construction. | + | [[File:FT1.png|250px|thumb|fig.3A:FT cDNA of Arabidopsis thaliana.]] |
| - | + | FT gene has 2 BioBrick restriction enzyme sites, EcoR1 and Pst1 (fig.3A.) So we did mutation to delete the sites before we start construction.<br> | |
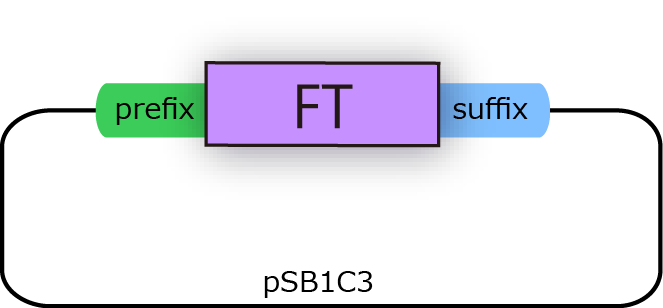
| - | We successfully cloned FT gene into BioBrick plasmid backbone, pSB1C3 with prefix and suffix(fig.3B.)<br> | + | We successfully cloned FT gene into BioBrick plasmid backbone, pSB1C3 with prefix and suffix (fig.3B.)<br> |
| - | [[File:FT2.png| | + | [[File:FT2.png|250px|thumb|fig.3B:FT BioBrick]] |
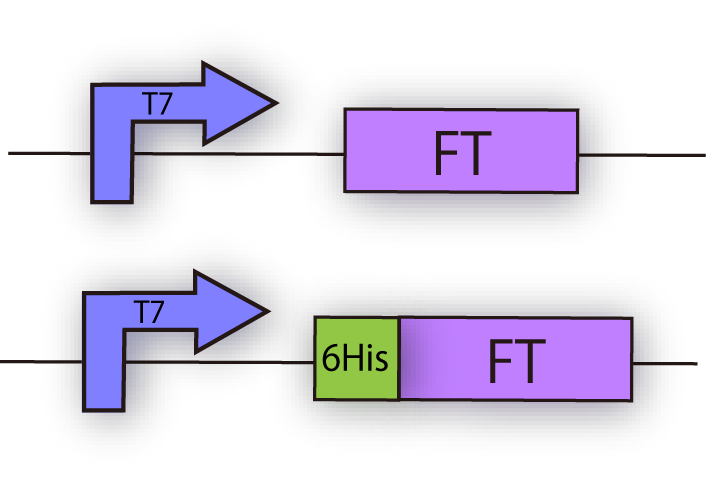
| - | We constructed the following plasmid to check the expression of FT by Western blotting, using FT specific antibody. FT and 6x his tagged FT are regulated by T7 promoter, BBa_I719005 and strong RBS, BBa_B0034. | + | We constructed the following plasmid to check the expression of FT by Western blotting, using FT specific antibody. FT and 6x his tagged FT are regulated by T7 promoter, BBa_I719005 and strong RBS, BBa_B0034 (fig.3C.) |
| - | <br>[[File:FTconstruction.png| | + | <br>[[File:FTconstruction.png|250px|thumb|fig.3C:FT construction]]<br><br><br><br><br><br><br><br><br><br><br><br><br> |
==Western blotting== | ==Western blotting== | ||
Revision as of 07:57, 24 September 2012
Contents |
Abstract
We will make "Flower Fairy E.coli" that makes and introduces florigen into plant cell and makes flowers bloom. Flower Fairy E.coli makes FLOWERING LOCUS T (Florigen), secretes florigen through TAT secretion system, and introduces florigen into plant cell using R9 peptide.
This florigen group try to confirm that E.coli can make florigen that acts correctly and that protein with R9 peptide can penetrate cell wall and membrane.
Introduction
What is Florigen?
It has always been a human dream to control the flowering of plants at will, and various studies have been done to realize this dream. Little was known about froligen, a plant hormone which promotes flowering for about seventy years since the existence of the hormone was suggested and it was named froligen, but recent studies have revealed more and more about it.
Froligen is a 20kDa of small protein composed of 175 amino acids which are encoded by the FLOWERING LOCUS T (FT) gene. The expression of the FT gene is promoted inside the leaf phloem companion cells by several specific conditions such as long photoperiods, vermalisation, age and so on. After transcripted, FT mRNA is translated into a protein and then transported to the shoot apex thorough sieve tubes. In the shoot apex, FT is combined with FD, which works as a transcriptional factor, and activates floral meristem identity genes such as APETALA 1 (AP1), FRUITFUL (FUL), and LEAFY (LFY). These genes play a significant role in the transformation of the form of a shoot meristem, converting it into a flower bud. Considering the effect of FT on flowering as mentioned above, we can say that E. coli with a capacity of producing FT are able to bloom flowers, as if they were flower fairies!
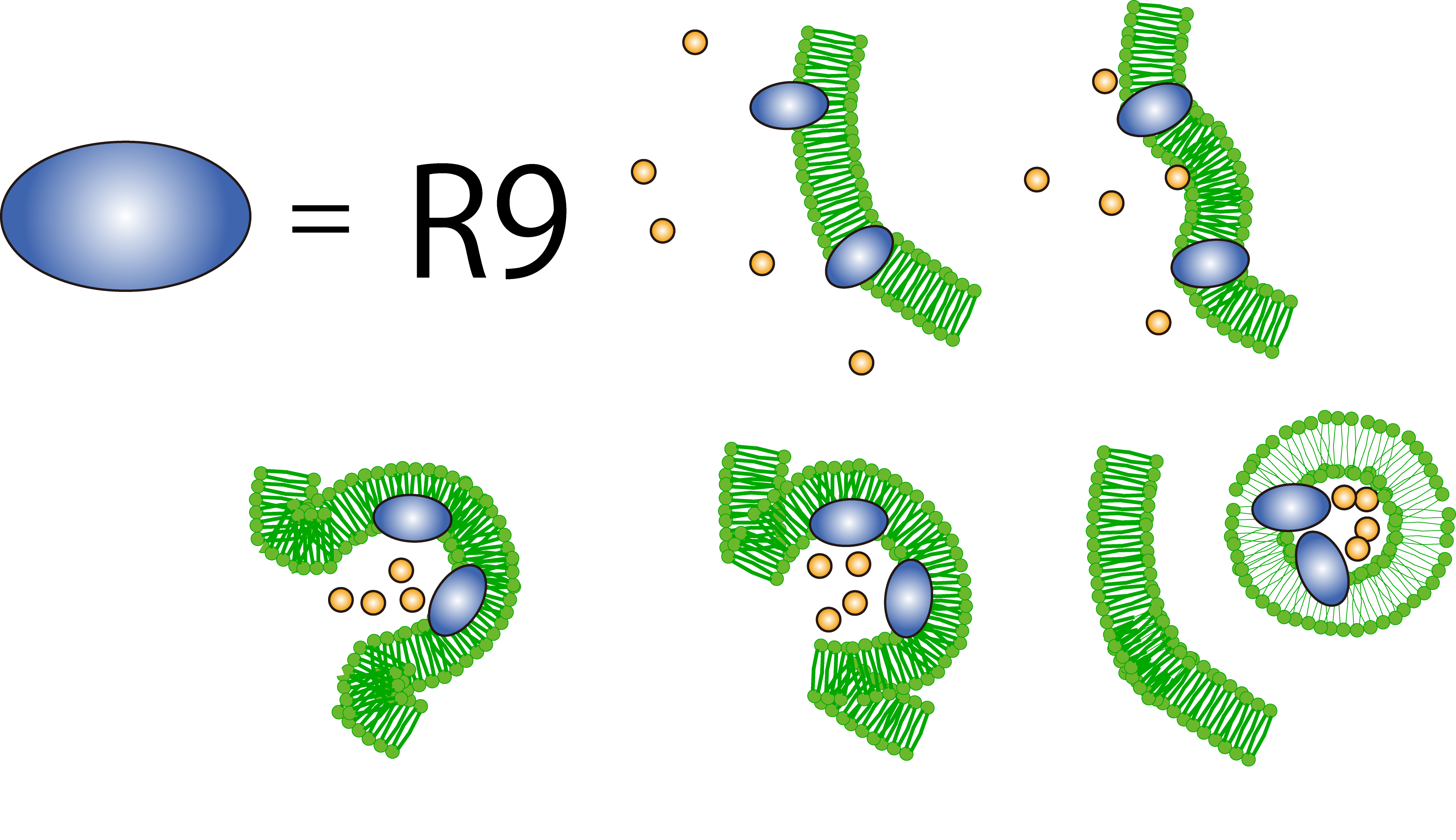
What is R9 peptide?
To realize flower fairy E. coli, FT secreted by E. coli must be absorbed in plant cells. We used polyarginine, a type of cell penetrating peptide (CPP), in order to transport FT protein into plant cells. The type is R9 peptide, which comprises a sequence conjugated to nine arginine residues.
Polyarginine peptide is thought to act on a cell membrane and cause a specific form of endocytosis, that is, macropinocytosis. Macropinocytosis is not caused by an invagination of the cell membrane, but by the growth on the actin mambrane from protrusions into vesicles called macropinosomes, and no receptors are necessary for the process. It is reported that plants use CPP to transport biomolecules such as proteins inside the cell, in spite of their cell walls.
Method
1. Expression of FT in E.coli
Construction
FT gene has 2 BioBrick restriction enzyme sites, EcoR1 and Pst1 (fig.3A.) So we did mutation to delete the sites before we start construction.
We successfully cloned FT gene into BioBrick plasmid backbone, pSB1C3 with prefix and suffix (fig.3B.)
We constructed the following plasmid to check the expression of FT by Western blotting, using FT specific antibody. FT and 6x his tagged FT are regulated by T7 promoter, BBa_I719005 and strong RBS, BBa_B0034 (fig.3C.)
Western blotting
1. Pre-experiment
First, we checked the specificity of anti GFP monoclonal antibody.
We characterized the existing GFP generator parts, [http://partsregistry.org/Part:BBa_I746915 BBa_I746915].
The parts is consist of T7 promoter 6-his tagged superfolder GFP.
Cells were precultured overnight and diluted into fresh SOC medium. IPTG was added when OD600 was approx. 0.7, then cells were incubated for 4h at 37°C. 100µL of culture was used for SDS-PAGE.
2. R9 peptide fusion GFP
Next, using following construction, we tried to check the expression of R9 peptide fusion GFP.
Cells were precultured overnight and diluted into fresh SOC medium. IPTG was added when OD600 was approx. 0.7, then cells were incubated for 4h at 37°C. 100µL of culture was used for SDS-PAGE.
3. GFP imaging assay for characterizing R9 peptide
Following plasmid was constructed to check the function of R9 peptide.
4. RT-PCR assay for characterizing FT
Result
1. Pre-experiment
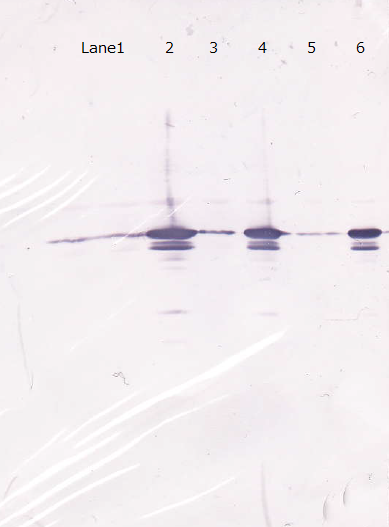
Following figure shows the result of western blotting of [http://partsregistry.org/Part:BBa_I746915 BBa_I746915].
Unfortunately, we used inappropriate molecular marker and could'nt confirm the molecular weights of samples.
Lane1 : IPTG 0mM, sample 10µL
Lane2 : IPTG 1mM, sample 10µL
Lane3 : IPTG 0mM, sample 5µL
Lane4 : IPTG 1mM, sample 5µL
Lane5 : IPTG 0mM, sample 2µL
Lane6 : IPTG 1mM, sample 2µL
2. R9 peptide fusion GFP
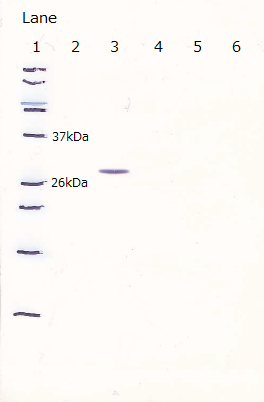
After the 4h of IPTG induction, we noticed that E.coli expressing R9::GFP were growing poorly.
Moreover, we couldn't get any bands of R9::GFP, as shown in the following figure.
Lane1 : Molecular marker
Lane2 : GFP([http://partsregistry.org/Part:BBa_I746915 BBa_I746915]) IPTG 0mM, sample 10µL
Lane3 : GFP([http://partsregistry.org/Part:BBa_I746915 BBa_I746915]) IPTG 1mM, sample 10µL
Lane4 : R9::GFP IPTG 0mM, sample 10µL
Lane5 : R9::GFP IPTG 1mM, sample 10µL
Discussion
Reference
[1]Microsugar Chang et al. (2005)"Cellular internalization of fluorescent proteins via arginine-rich intracellular delivery peptide in plant cells" Plant Cell Physiol, 46(3), 482–488
[2]Paula Teper-Bamnolker and Alon Samach1 (2005) "The flowering integrator FT regulates SEPALLATA3 and
FRUITFULL accumulation in Arabidopsis leaves" The Plant Cell, 17, 2661–2675
[3]Philip A. Wigge et al. "Integration of spatial and temporal information during floral induction in Arabidopsis
[4]Sara Trabulo et al.(2010). "Cell-penetrating peptides—mechanisms of cellular uptake and generation of delivery systems" Pharmaceuticals, 3, 961-993
 "
"