Team:Amsterdam/modeling/generaldesign
From 2012.igem.org
(Difference between revisions)
(→Model definition) |
(→Model definition) |
||
| Line 128: | Line 128: | ||
<center> | <center> | ||
| - | [[File:Timelapse.jpg|image]] | + | [[File:Timelapse.jpg|image]]<br> |
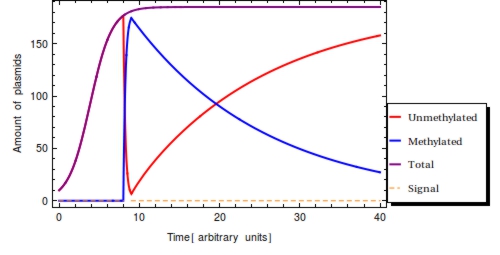
<b>Figure 2</b>. Time simulation of the system of ODE’s. Input signal <math>S(t)</math> with <math>s_{\text{on}} = 3</math> and <math>s_{{\text{off}}} = 4</math>. Detection of the signal converts all <math>P_{0}</math> (red) to <math>P_{1}</math> (blue) on a short time scale. After the amount <math>P_{1}</math> will start to diminish due to cell division. Eventually, the steady state will be restored once again and the cell’s capacity for plasmids will be completely taken up by <math>P_{0}</math> plasmids. | <b>Figure 2</b>. Time simulation of the system of ODE’s. Input signal <math>S(t)</math> with <math>s_{\text{on}} = 3</math> and <math>s_{{\text{off}}} = 4</math>. Detection of the signal converts all <math>P_{0}</math> (red) to <math>P_{1}</math> (blue) on a short time scale. After the amount <math>P_{1}</math> will start to diminish due to cell division. Eventually, the steady state will be restored once again and the cell’s capacity for plasmids will be completely taken up by <math>P_{0}</math> plasmids. | ||
</center> | </center> | ||
| - | Unknown variables affecting <math>F(t)</math> in a real-life setting would be the time of signal onset, signal duration and signal strength. Knowing the values for two of these three values, the value of the third can be solved for. Here we will simply assume maximal signal strength | + | Unknown variables affecting <math>F(t)</math> in a real-life setting would be the time of signal onset, signal duration and signal strength. Knowing the values for two of these three values, the value of the third can be solved for. Here we will simply assume maximal signal strength during <math>s_{\text{on}}</math> and <math>s_{\text{off}}</math>. |
The response rate <math>\omega</math> of the ''Cellular Logbook'' could limit <math>F(t)</math>, as a low <math>\omega</math> might yield incomplete methylation of all plasmids before <math>\s_{\text{off}}</math>. This rate should be experimentally determinable before actual deployment and application of our system and is more closely looked at in the next section. It is likely to be several magnitudes greater than the cellular division rate, however. Every single gene on a plasmid is thus expected to be methylated within at most 5 minutes of registering of the signal. | The response rate <math>\omega</math> of the ''Cellular Logbook'' could limit <math>F(t)</math>, as a low <math>\omega</math> might yield incomplete methylation of all plasmids before <math>\s_{\text{off}}</math>. This rate should be experimentally determinable before actual deployment and application of our system and is more closely looked at in the next section. It is likely to be several magnitudes greater than the cellular division rate, however. Every single gene on a plasmid is thus expected to be methylated within at most 5 minutes of registering of the signal. | ||
Revision as of 06:39, 21 September 2012
 "
"