Template:Kyoto/Project/FlowerFairy
From 2012.igem.org
(→2.SECRETION) |
(→We improved usability of TorA signal biobrick) |
||
| Line 96: | Line 96: | ||
To solve these problems, we produced new applicable TorA signal [http://partsregistry.org/Part:BBa_K797002 BBa_K797002] (Fig.2-2). Our part has two improvements. [http://partsregistry.org/Part:BBa_K797002 BBa_K797002] contains RBS and indels to prevent the emergence of stop codon between signal region and target cording sequence. We sequenced [http://partsregistry.org/Part:BBa_K797002 BBa_K797002] and confirmed that stop codon did not appear when we used it in Standard and 3A assembly. Using green fluorescent protein (GFP) as a target protein, we constructed plasmid of LacP-RBS-TorA-GFP-DT and introduced it into E.coli. After that, we observed the TorA-GFP-fusion-expressing cells (Fig.2-3) suggesting that the TorA-GFP fusion was successfully expressed. This meant RBS in our TorA signal worked well and stop codon didn't appear after assembly. <br> | To solve these problems, we produced new applicable TorA signal [http://partsregistry.org/Part:BBa_K797002 BBa_K797002] (Fig.2-2). Our part has two improvements. [http://partsregistry.org/Part:BBa_K797002 BBa_K797002] contains RBS and indels to prevent the emergence of stop codon between signal region and target cording sequence. We sequenced [http://partsregistry.org/Part:BBa_K797002 BBa_K797002] and confirmed that stop codon did not appear when we used it in Standard and 3A assembly. Using green fluorescent protein (GFP) as a target protein, we constructed plasmid of LacP-RBS-TorA-GFP-DT and introduced it into E.coli. After that, we observed the TorA-GFP-fusion-expressing cells (Fig.2-3) suggesting that the TorA-GFP fusion was successfully expressed. This meant RBS in our TorA signal worked well and stop codon didn't appear after assembly. <br> | ||
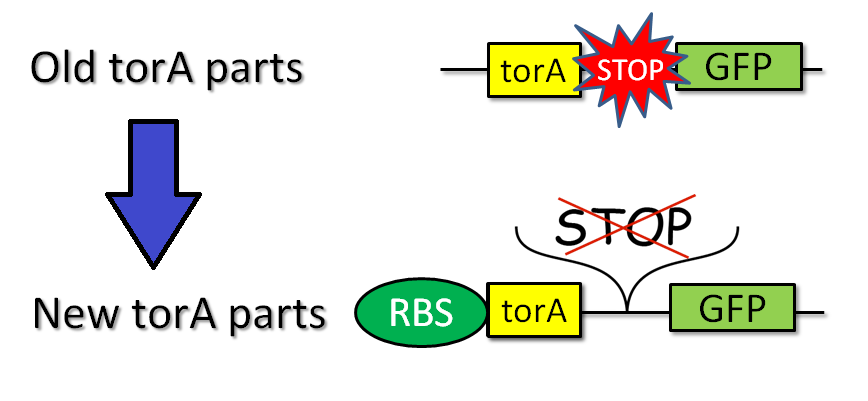
| - | [[File:Sec-inner-torA2.png|thumb|center|600px|Fig.2-3 | + | [[File:Sec-inner-torA2.png|thumb|center|600px|Fig.2-3 Previous TorA signal biobricks (such as BBa_K638402) and our modified TorA signal (BBa_K797002)]] |
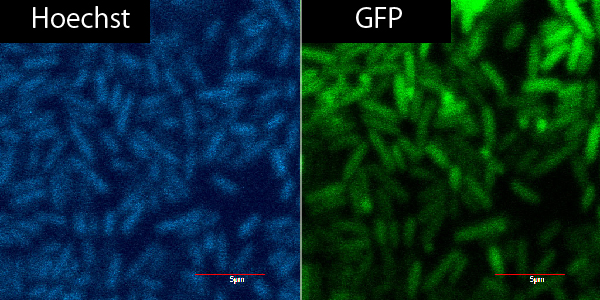
| - | [[File:光る大腸菌.jpg|thumb|center|600px|Fig.2-4 ''E.coli'' (left) sample stained with Hoechst and observed with 352nm wavelength. ''E.coli'' (right) sample having Lacp-TorA-GFP-DT and observed with 489nm wavelength.]] | + | [[File:光る大腸菌.jpg|thumb|center|600px|Fig.2-4 ''E.coli'' (left) sample stained with Hoechst and observed with 352nm wavelength. Hoechst can stain DNA. ''E.coli'' (right) sample having Lacp-TorA-GFP-DT and observed with 489nm wavelength.]] |
====Kil protein enables proteins to go through outer membrane==== | ====Kil protein enables proteins to go through outer membrane==== | ||
Revision as of 17:31, 26 October 2012
Realizing Flower Fairy in real world
Have you ever seen flower fairies? Probably the answer is, "no" (though some of you might have come across them in your childhood), because they are imaginary creatures which exist only in fairy tales. Don’t you think it would be wonderful if you could live with flower fairies? In addition to the happy feelings, their lovely power to make flowers bloom would be profitable for us in many ways, such as application to agriculture. That is why we have set our project for realizing Flower Fairy E.coli with synthetic biology!!
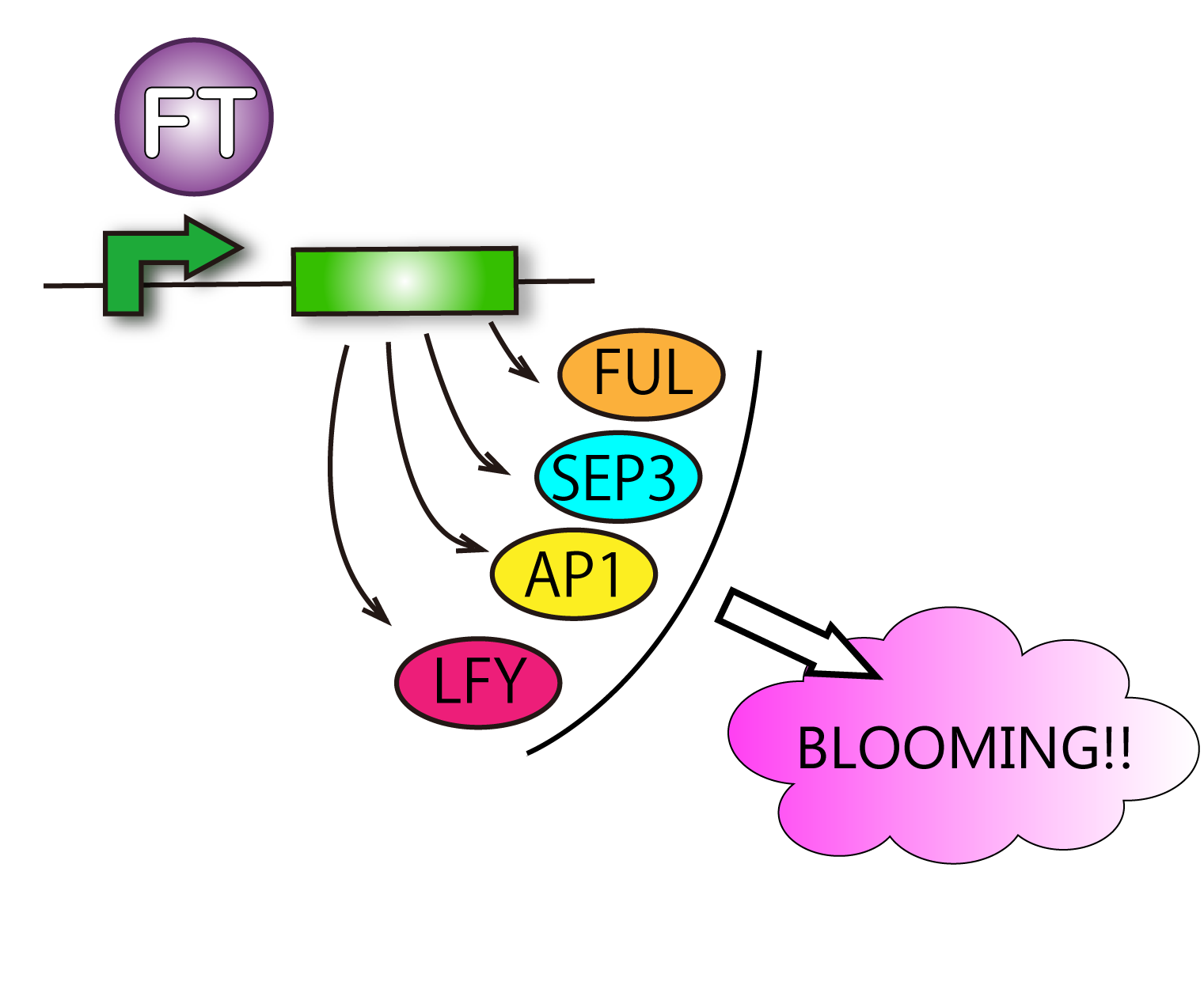
Our goal is to produce E.coli which can make flowers bloom as Flower Fairies. To make it possible, we focused on FT protein, known as Florigen.This protein is a kind of plant hormone. First, FT protein is produced in leaves, and then move to the shoot apex and bloom flowers. Therefore, FT protein is the key to our project.
Our Goal is to induce flower formation just by putting Flower Fairy E.coli on leaves!
When you want to use our Flower Fairy E.coli, all you have to do is just put them on plant leaves! When you spread Flower Fairy E.coli, FT protein is secreted and penetrate the cell membrane of a plant, and the plant starts blooming.
We had to go through four steps in order to achieve our goal――Flower Fairy E.coli.
These four steps are “EXPRESSION”,”SECRETION”,
”PENETRATION”, and ”ACTIVATION”
In each step, we have some problems to be attacked.
“EXPRESSION”; It is unclear whether E.coli (prokaryote) can express FT protein, because FT protein is derived from plant cells (eukaryote).
”SECRETION”; After produced, FT protein has to get out of the E.coli.
”PENETRATION”; FT protein has to penetrate into plant cells.
”ACTIVATION”; Even if FT protein could get into the cells, it is not clear whether FT protein produced by E.coli can activate genes in shoot apex cells and induce flower formation.
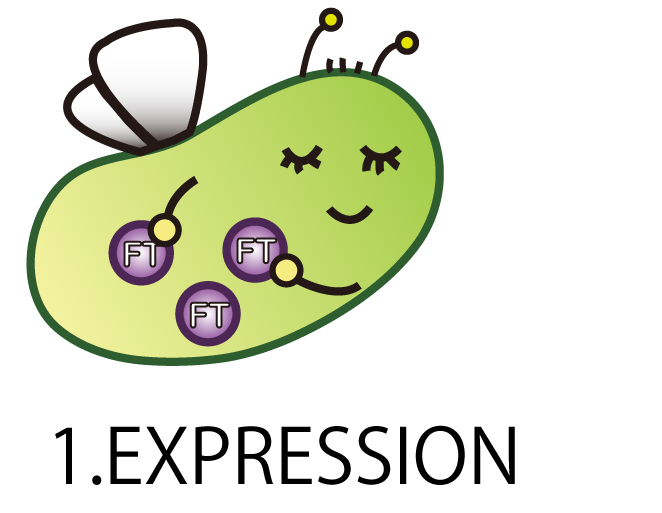
1.EXPRESSION
The first step is EXPRESSION.
We needed to enable E.coli to produce FT protein.
As a matter of course, E.coli doesn’t have FT gene. Therefore we had to make a new BioBrick part of FT gene and introduce it into E.coli.
Modifying FT gene for Biobrick
FT gene is derived from Arabidopsis thaliana, a model plant. Professor Araki in Kyoto University kindly gave us FT cDNA in TOPO blunt end 2(Invitrogen). FT cDNA has two cleavage sites of iGEM restriction enzymes, EcoR1 and Pst1 (Fig.1-1 A), therefore we modified FT gene sequence by Inverse PCR with primers containing two base mismatches between primer and cDNA(Fig.1-1 B). As a result, we could get mutated FT cDNA, which are not cleaved by iGEM restriction enzymes, and then we added prefix and suffix to FT. Finally, we could make new biobrick parts of FT (Fig.1-1 C).
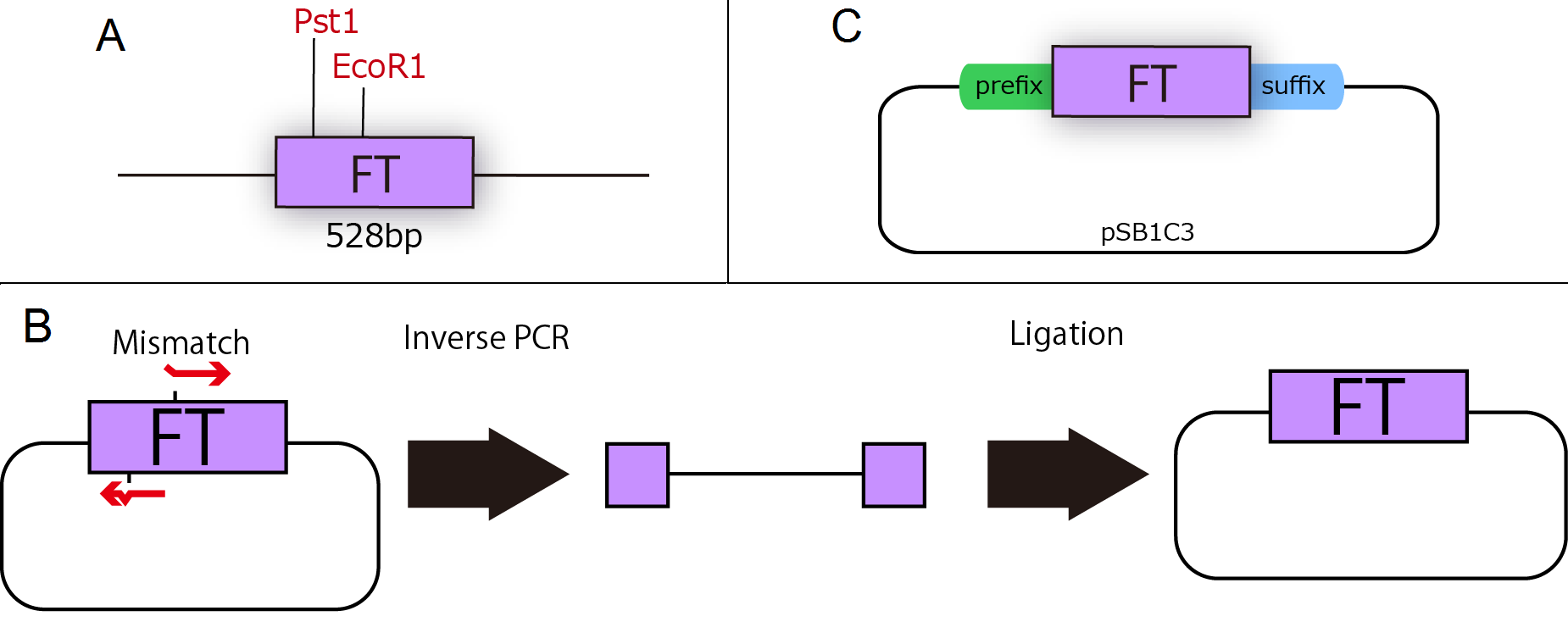
A : Necessity for mutation of FT cDNA. FT sequence had two cleavage sites of iGEM restriction enzymes.
B : Inverse PCR Method
Inverse PCR is a measure of mutating FT gene segment. Inverse direction of primers and designing those including mismatch residue leads to mutation of the original plasmid.
C : Standardization of FT cDNA. We had mutated and added prefix and suffix to FT.
Confirming expression of FT
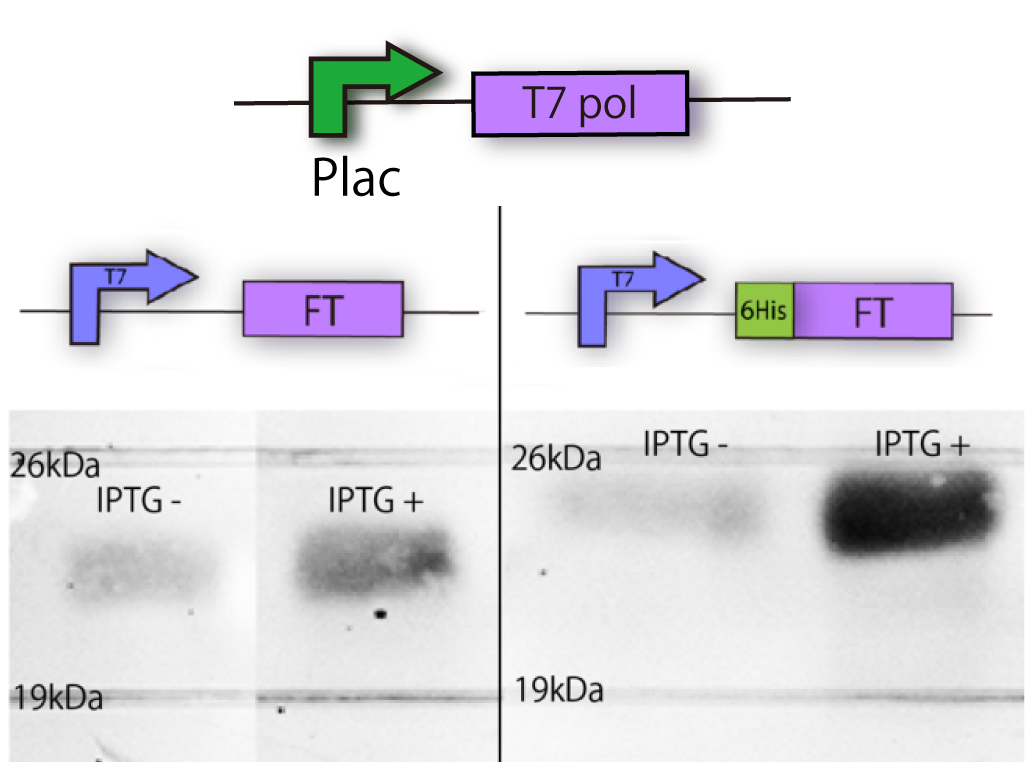
We constructed the plasmid shown in the Fig.1-2. One was FT gene with T7 promoter ([http://partsregistry.org/Part:BBa_I719005 BBa_I719005]) and 6His tag, and the other was FT gene only with T7 promoter. T7 promoter is a strong promotor regulated by T7polymerase. In our strain of E.coli, the expression of T7 pol can be induced by IPTG. 6His tag, which is used in later steps, enabled us to purify FT protein from E.coli with affinity chromatography.
We needed to confirm that E. coli can express FT protein, because there was a possibility that E.coli can not translate FT or FT is unstable or toxic in E.coli.
In order to confirm the expression of FT protein, we performed Western blotting using anti-FT antibody.
As a result, FT and 6 His:FT bands were observed at the expected molecular weight region(Fig.1-2).
We succeeded in making mutation and confirming the expression of FT and 6 His:FT proteins in E.coli!
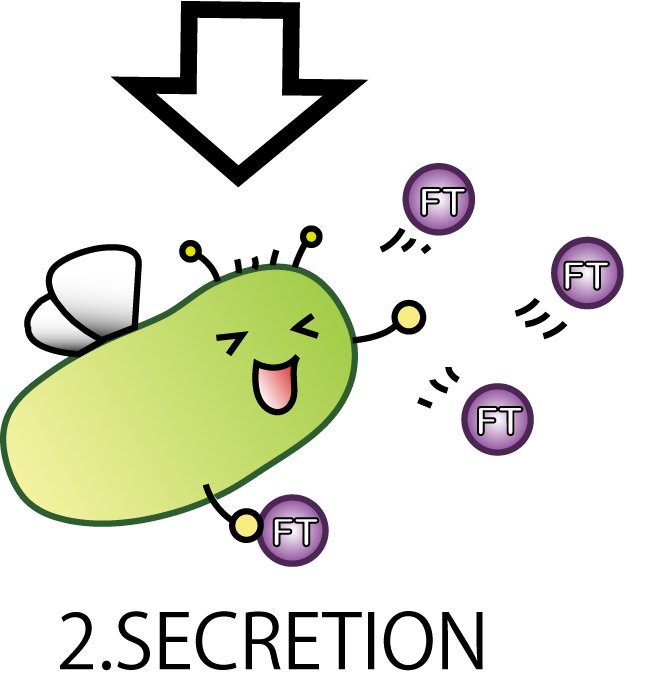
2.SECRETION
The second step is SECRETION. We want E.coli to secrete FT protein to outside of the cell.
Now our E.coli can produce FT protein, but a big issue still remains:
how they can transport proteins to the outside of the cells? Many people might think cell lysis is the best way.
But in our project lysis is not very good, because it would possibly cause all fairies' deaths. We hoped to see the continuous effect of Flower Fairy E.coli, not just temporary. So, we adopted a method other than lysis which enables E.coli to transport FT protein continuously.
Proteins are required to go through two membranes
E.coli have two membranes: inner membrane and outer membrane. To transport FT protein to outside of E.coli, therefore, FT protein has to pass through the two membranes. So we searched for secretion systems to make FT proteins go through these membranes.
TorA signal enables proteins to go through the inner membrane via tat pathway
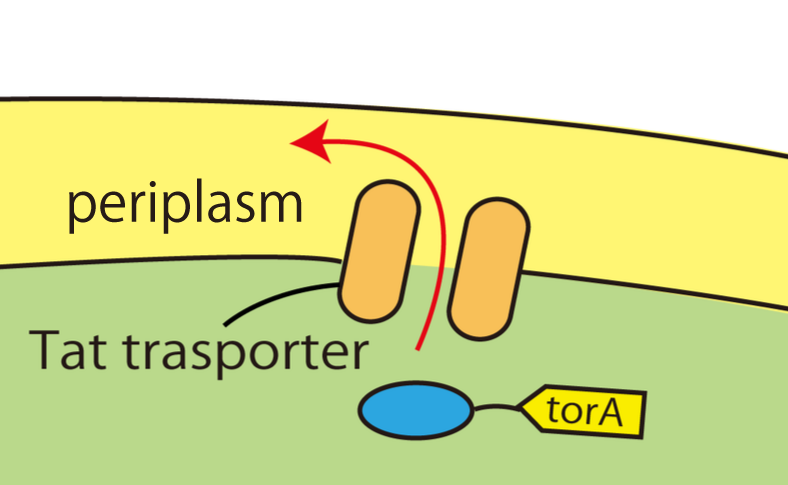
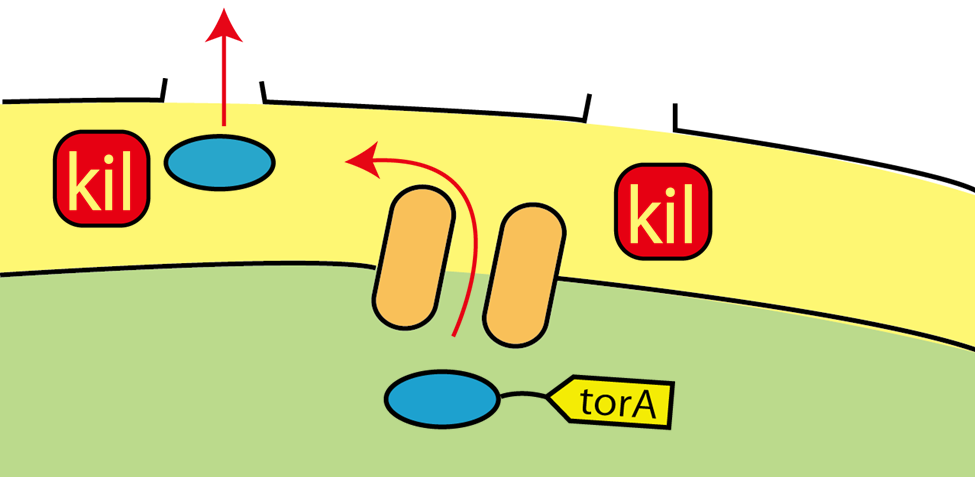
Wild E.coli have many secretion systems. In order to enable proteins to go through inner membrane, we decided to use one of these systems, called Twin Arginate Translocation(Tat) pathway. Tat pathway is more suitable than other secretion systems for us, because by Tat pathway E.coli secrete proteins into the periplasm keeping proteins’ conformation and function. The following is the mechanism of Tat pathway; a Tat transporter recognizes TorA signal, and transports into the periplasm only proteins that have a TorA signal at their N terminals.
We improved usability of TorA signal biobrick
We tried to combine TorA signal with FT protein. Searching previous iGEM projects, we actually found TorA signals as iGEM parts (such as [http://partsregistry.org/Part:BBa_K638402 BBa_K638402]) submitted by other teams. These parts, however, have two big problems. One problem is that these parts do not have RBS, which regulate additional processing time for iGEMers who use the parts. The other problem is that stop codon appear between signal region and target coding sequence when these parts are combined with some other parts by standard or 3A assembly. In short, therefore, when iGEMers use these parts, TorA-fusion protein would not be expressed at all, but TorA would be expressed.
To solve these problems, we produced new applicable TorA signal [http://partsregistry.org/Part:BBa_K797002 BBa_K797002] (Fig.2-2). Our part has two improvements. [http://partsregistry.org/Part:BBa_K797002 BBa_K797002] contains RBS and indels to prevent the emergence of stop codon between signal region and target cording sequence. We sequenced [http://partsregistry.org/Part:BBa_K797002 BBa_K797002] and confirmed that stop codon did not appear when we used it in Standard and 3A assembly. Using green fluorescent protein (GFP) as a target protein, we constructed plasmid of LacP-RBS-TorA-GFP-DT and introduced it into E.coli. After that, we observed the TorA-GFP-fusion-expressing cells (Fig.2-3) suggesting that the TorA-GFP fusion was successfully expressed. This meant RBS in our TorA signal worked well and stop codon didn't appear after assembly.
Kil protein enables proteins to go through outer membrane
Using Tat secretion pathway, FT protein is transported into periplasm. Next, FT protein needs to be transported out of E.coli. For this purpose, we used kil protein, which is derived from λ phage. Kil protein makes holes in outer membrane of E.coli. So on our project, we introduced kil gene into E.coli and want FT protein to go through these holes. But the function of outer membrane is essential for E.coli to survive. Overexpression of kil gene, therefore, causes cell death. For this reason, we must check whether kil gene is harmful or not under our condition.
Kil protein has no effect on E.coli's death
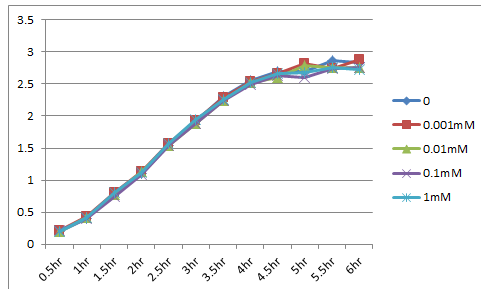
We made the construction lacp-RBS-kil-double terminator, whose backbone is pSB3C5. After culturing for 18hr at 37℃, we eliminated the supernatant using a centrifuge, and diluted it until OD600=0.1. Then we dispensed it. The dispense volume was 3mL. We added 0/0.001/0.01/0.1/1mM IPTG to each. While culturing again at 37℃, we measured OD600. The figure below shows the results. These results indicated that the expression of LacP-RBS-kil-DT (pSB3C5) makes no effect on the survival of E.coli.
We established the base of secretion system. When we apply these systems, tat pathway and kil protein, to Flower Fairy E.coli, we can make E.coli to secrete FT protein without cell lysis!!(Fig.2-7)
Improvement of secretion system
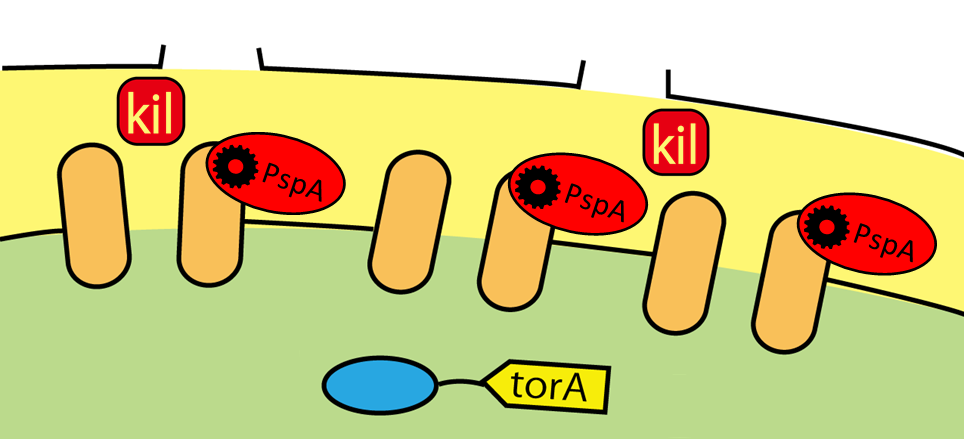
Now we can make E.coli secrete FT proteins outside of them by using Tat pathway and kil protein. But E. coli secrete only a small amount of FT proteins when they use Tat transporters they originally have. To make E.coli secrete enough amount of FT proteins, we needed to improve the efficiency of their secretion systems. To reach this goal, we used two genes. One is composed of TatA, TatB, and TatC, which makes Tat transporter. Another is phage-shock protein A (pspA), which wild E.coli have. When their inner membrane is damaged, pspA gene is expressed ,and pspA maintains membrane potential and H+ concentration gradient between periplasm and cytoplasm. On our project, we expected that E.coli would secrete more proteins if those two genes are expressed more. It is because in our plan inner membrane and outer membrane have many holes and E.coli are exposed to the membrane stress. We, therefore, constructed Tat secretion cassette with constitutive promoter [http://partsregistry.org/Part:BBa_K797004 (BBa_K797004)].(Fig.2-8)
This part includes TatA, B and C proteins coding region and pspA. By using this part, the amount of Tat transporter is increased and pspA diminishes stress of membranes. In short, we can make E.coli secrete more proteins with TorA.(Fig.2-9) <
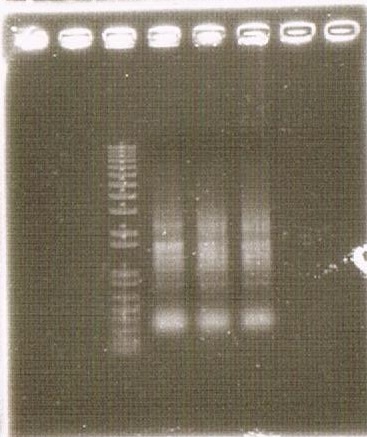
Kyoto 2012 suggests this new way of secretion, that is using Tat pathway and kil protein, and provides iGEMers with this cassette regulated by constitutive promoter. We checked the sequence of TatABC [http://partsregistry.org/Part:BBa_K797000 (BBa_K797000)] and the sequence of pspA [http://partsregistry.org/Part:BBa_K797001 (BBa_K797001)] individually, and then, we made Tat construction composed of constitutive promoter [http://partsregistry.org/Part:BBa_J23107 (BBa_J23107)], TatABC [http://partsregistry.org/Part:BBa_K797000 (BBa_K797000)], pspA [http://partsregistry.org/Part:BBa_K797001 (BBa_K797001)] and double terminator [http://partsregistry.org/Part:BBa_B0015 (BBa_B0015)]. This Tat secretion cassette is too long device to sequence, so we performed electrophoresis of this cassette and confirmed the length of our parts.
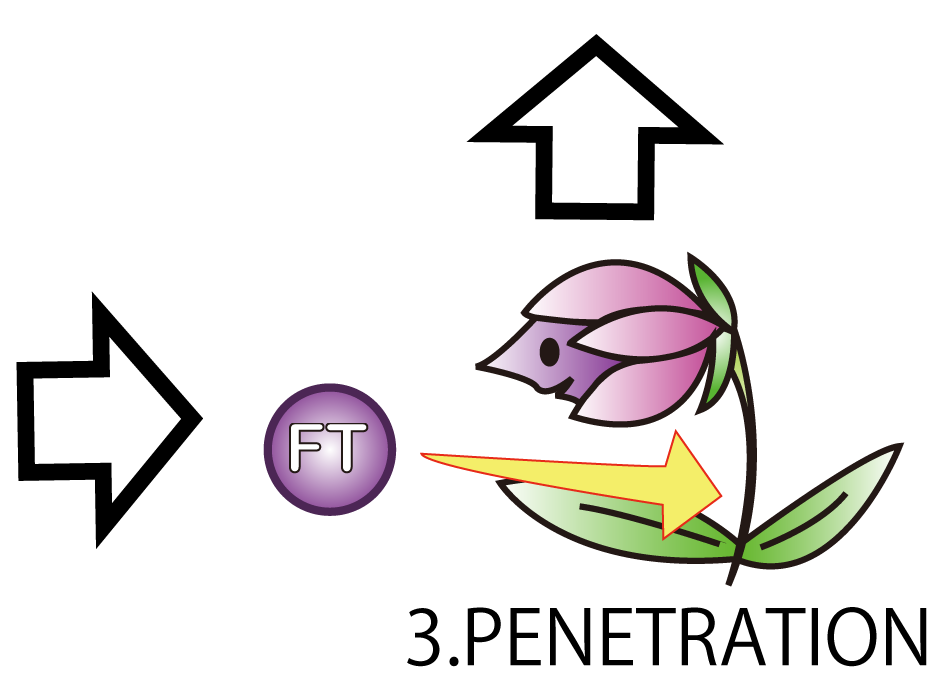
3.PENETRATION

The third step is PENETRATION. In order to induce flower formation, FT proteins from E.coli must enter into plant cells. This is because FT proteins upregulate other proteins which lead to flower formation in plant cells.
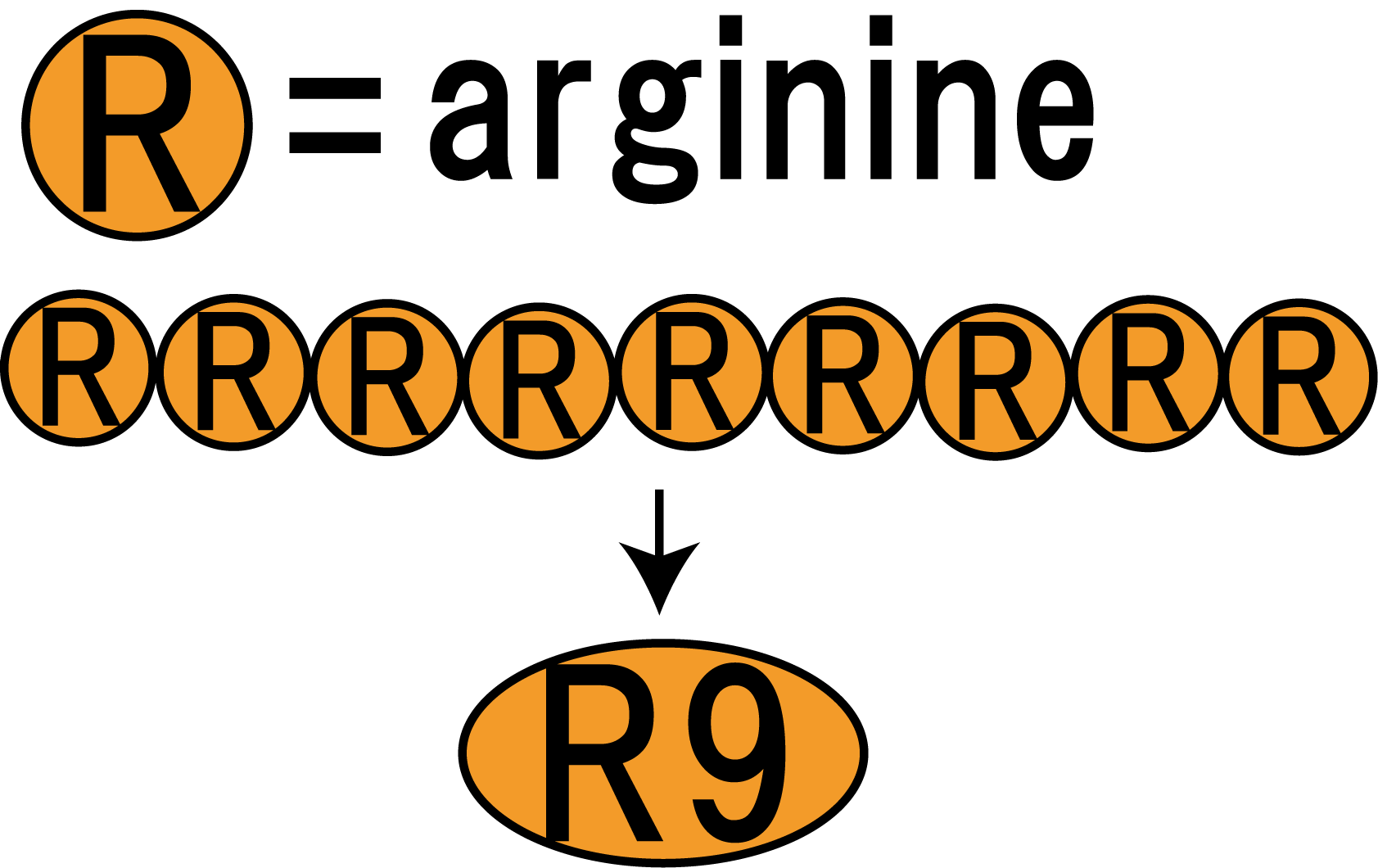
However, normally proteins cannot penetrate a cell membrane of a plant. Therefore, we needed a method to send FT protein into plant cells. Thanks to the advice from Doctor Washida, we found a method for the penetration of cell membrane. In this method, we use polyarginines called R9 peptides.
R9 peptide enables FT protein penetrate membranes by endocytosis
R9 peptide consists of nine arginine residues(Fig.3-1). It is known as a kind of CPP (Cell Penetrating Peptide). Arginine-rich peptides induce macropinocytos, a type of endocytosis. From this, we judged that R9 peptides were suitable for cell membrane penetration.
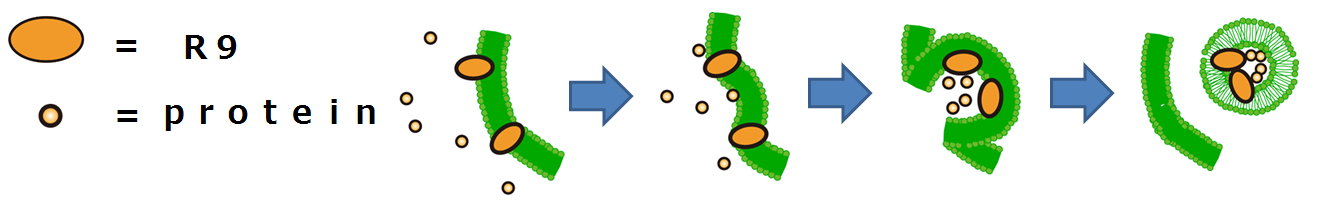
This is the mechanism of how R9 peptides work(Fig3-2).
Firstly, R9 peptides adhere to a cell membrane of a plant because of their hydrophobic character.
Secondly, the cell responds to this stimulus and starts to endocytose.
Finally, proteins around the endocytosing region of the cell is taken into the cell.
R9 peptides seem to work effectively regardless of whether or not R9 peptides are fused with target proteins.
However, there are few examples about endocytosis of plants, so we needed to check the function of R9 when used on plant cells.
From the R9 system, it induces endocytosis whether R9 peptides and target proteins are fused or not.
Considering this system, target proteins near the R9 peptides are taken by endocytosis.
In other words, it can be said that it induces endocytosis regardless of whichever R9 peptides and target proteins are fused.
We tried to wonder whether we should fuse R9 and target proteins.
From this system, we thought the shorter the distance between R9 peptides and target proteins is, the more easily they are taken into a plant cell. Therefore, we considered that fusing R9 peptide with a target protein may leads to higher efficiency.
So, we tried to fuse R9 peptides with target proteins to increase penetration efficiency
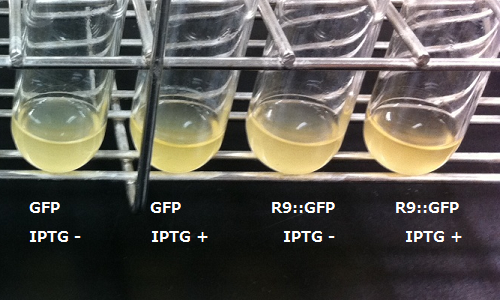
We transformed E.coli, arranging R9's sequence and target protein's gene in line. And after that we tried to make E.coli express R9::GFP fusion protein.
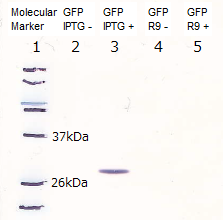
In order to visualize the function of R9, we tried to prepare R9::GFP fusion protein. However, E.coli expressing the R9::GFP fusion protein was poor in growth(Fig3-3). Then, we checked R9's effect on R9::GFP fusion protein's expression by Western blotting and RT-PCR. We succeeded in confirming the existence of the mRNA (Fig3-4), but we did not find the protein (Fig3-5). These results indicated poor translation or quick breakdown of the protein.
We used the existing GFP generator part, [http://partsregistry.org/Part:BBa_I746915 BBa_I746915].
Fig.3-4 Western blotting for checking the expression. No band of R9::GFP fusion protein. |
We prepared R9 and GFP separately and to check the R9 peptides function
We wondered whether R9 system really works properly and puts FT proteins into plant cells. Although animal cells often endocytose, there are few examples of plant cell’s endocytosis. Therefore, we need to verify the system. To verify it, we performed following experiment(Fig3-6).
First, we removed the cuticle of Arabidopsis thaliana's leaves by scratching them. Arabidopsis thaliana is a model plant. Second, we soaked the leaves into a solution of only GFP, or GFP and R9 for each. These GFP proteins were purified with 6 His tag inserted in our 1st Step, Expression. After 5 minutes we washed cells by PBS in order to wash GFP and R9 peptide away from leaves. After that, we compared leaves including only GFP and leaves including R9 and GFP for each.
Then we succeeded in getting the Figure of GFP fluorescence(Fig3-7).
The control groups on the left were soaked in only GFP, and the experimental group on the right were soaked in GFP and R9. These fluorescence indicated that R9 peptides work properly kept GFP in or around plant cells. This Figure strongly suggests that R9 peptide works successfully and penetrates cell membrane with GFP, because this was taken by a confocal microscopy and seen as cross sections.
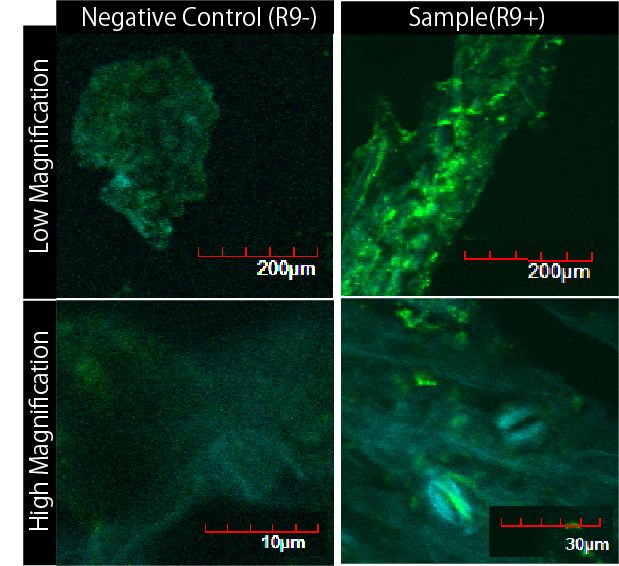
We can make FT protein penetrate cell membrane of plants by R9 peptide function!.
4. Activation

On the final step; Activation. We verified whether FT protein made in E.coli worked normally in plant cells.
Actually, FT protein is a transcriptional factor so that we can check the ability of FT protein by observing the transcript levels of genes FT protein upregulate.
The detail function of FT as a transcriptional factor
FT protein increases transcriptive activity of several proteins which lead to flower formation. For that reason it can be said that we have verified FT function when we have found rises of activities of the proteins.
Although such proteins activated by FT are various, we check FRUITFULL(FUL) and SEPALLATA3(SEP3). This is because SEP3 and FUL are activated in leaves. It is difficult for us to handle cells of tops of stems. So we focused on cells of leaves. Leaves' cells are easy to handle for us. We performed RT-PCR in order to investigate FT protein's function.
leave is suitable for checking the function of FT
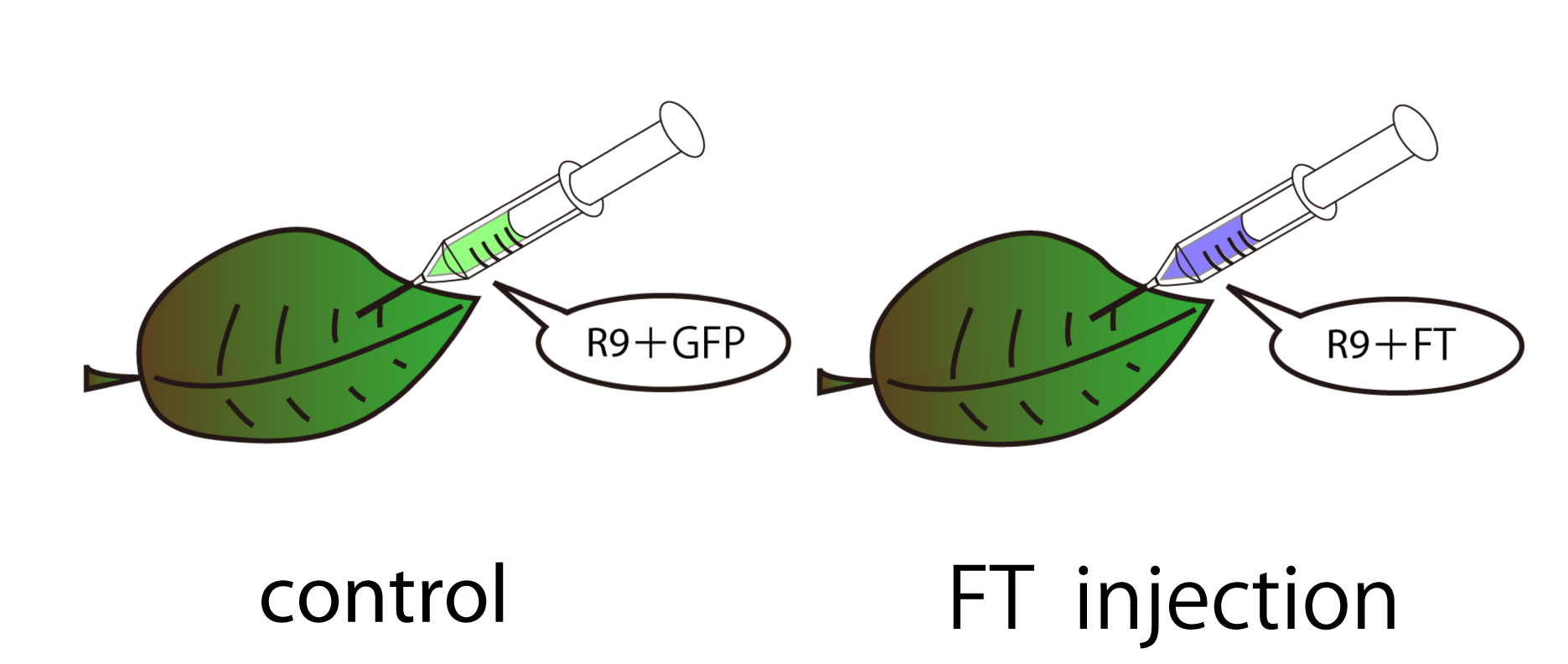
Injecting FT and verifying its function by RT-PCR
FT protein is injected by a syringe. Two types of samples were prepared, one is treated with FT and the other is treated with GFP as a control. GFP was used as a control to confirm the result of RT-PCR was derived from FT protein, not protein injection itself. GFP is suitable for control experiments because GFP's molecular weight(27kDa) is relatively similar to that of FT(20kDa.)
We performed RT-PCR to compare mRNA expression of Arabidopsis leaves treated with/without FT.
Fig.4-2 is the result of RT-PCR.

30mg of leaves of Arabidopsis thaliana before bolting were used for one sample. FT or GFP protein and R9 peptide were diluted in PBS (pH7.4), 50ug/L and 500ug/uL each. Leaves were soaked into FT-R9 or GFP-R9 solution for 5min. and incubated for 16hr. in PBS(pH7.4.) After incubation, leaves were freezed with liquid nitrogen and glinded immediately.
Total RNA was extracted by phenol-chloroform extraction. cDNA was synthesized by reverse transcription and used as templates of RT-PCR. TUBULIN was used for internal control of mRNA expression.
Lane1: TUBULIN (GFP-R9 treated) amplicon 61bp
Lane2: TUBULIN (FT-R9 treated)
Lane3: FUL (GFP-R9 treated) amplicon 132bp
Lane4: FUL (FT-R9 treated)
Lane5: SEP3 (GFP-R9 treated) amplicon 87bp
Lane6: SEP3 (FT-R9 treated)
Lane7: AP1 (GFP-R9 treated) amplicon 958bp
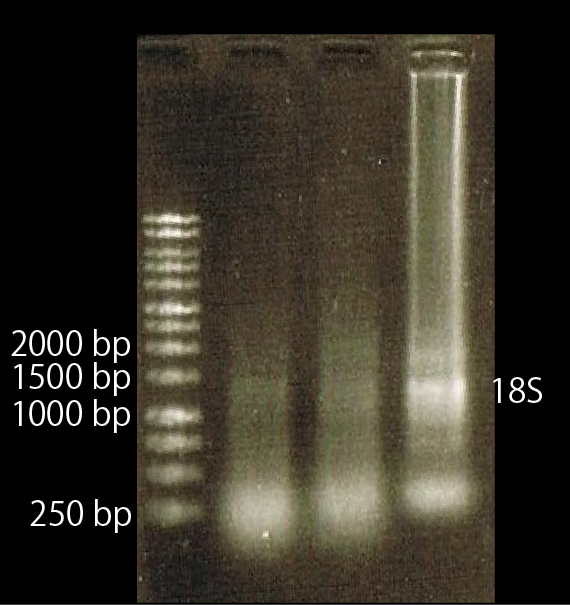
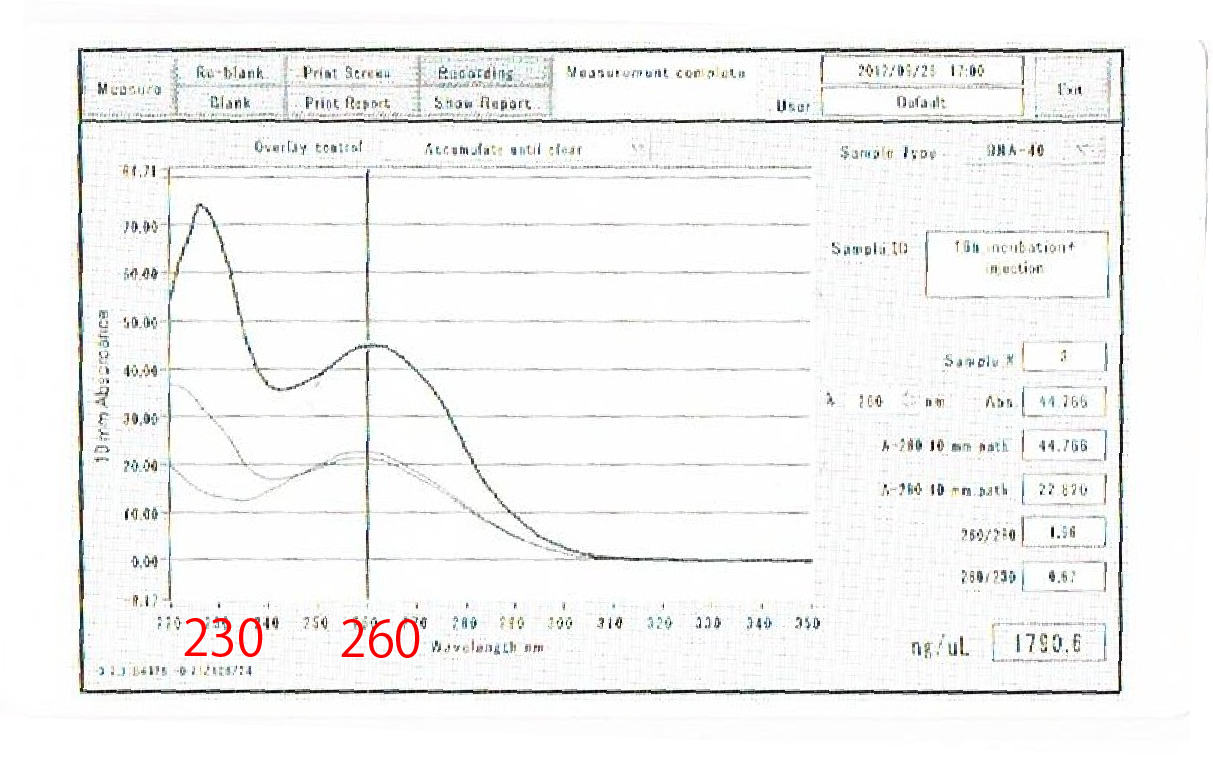
As shown above, we got no correct ampicon band, even of tubulin, which is high expressed gene we used as internal control. One possible reason of this failure is the poor quality of extracted RNA in this experiment. To check this, we compared the total RNA by electrophoresis, shown in fig.4-3.
As shown in the Fig.4-3, RNA samples in this time are degradated.
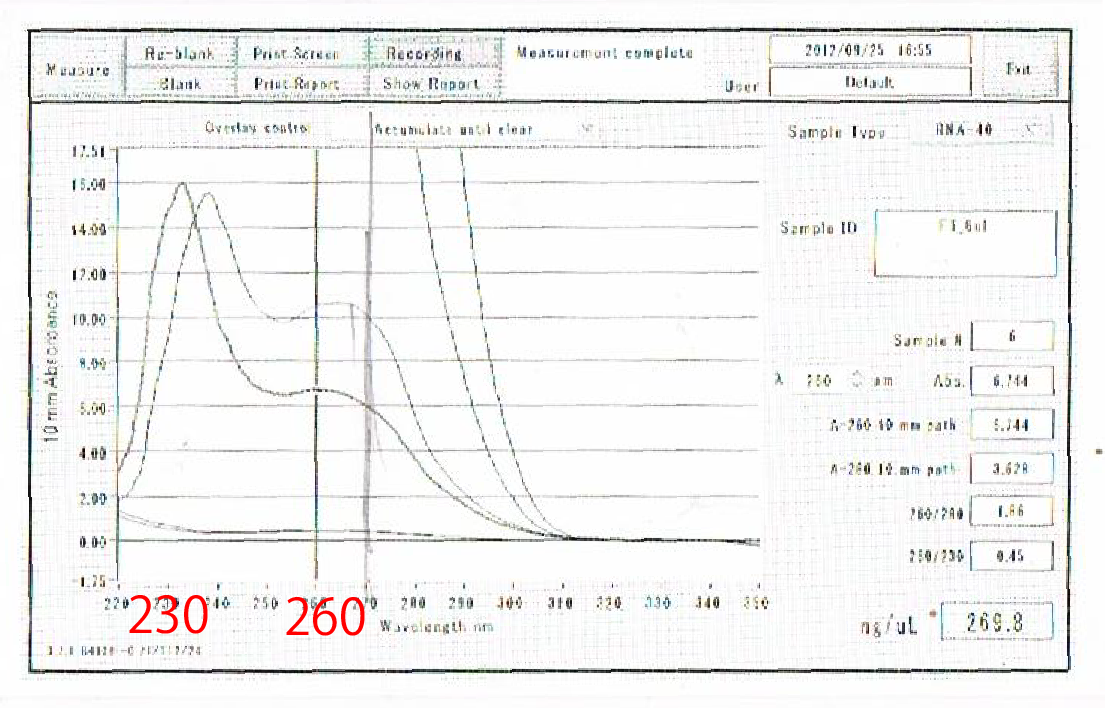
Moreover, the waveforms of them had a law peak at 260nm(Fig.4-4.)
So, it was required to improve the method of RNA extraction.
We improved following things;
1. Total leaf volume was increased.
2. Samples ware freezed with liquid nitrogen and suspended in ISOGEN more rapidly.
3. We centrifuged samples and collected supernatant twice after adding ISOGEN.
After the improvement, 260nm peak became higher and the degradation is minimized(Fig.4-4, 4-5.)
Then we used better quality of RNA for reverse transcription and retried RT-PCR.
Fig.4-6 shows the result of this RT-PCR.
improving the way of RNA extraction
conducting qPCR
Whether FT protein went into plant cell?
From these results, we could not confirm the function of FT, upregulation of FUL, SEP3, and AP1.
However, now that we can get good quality of RNA and amplify genes successfully, we are trying to check the function of FT.
Achievement
1.Expression
- Mutate FT sequence
- Standardize FT as an iGEM part
- Confirm expression of FT protein in E.coli
2.Secretion
- Modify TorA signal to be easy to use the signal more
- Construct Tat secretion cassette that contains Constitutive promoter, RBS, TatABCD, pspA, double terminator
- Standardize kil gene
- Multiply TatABC in order to strengthen Tat secretion system--not yet
3.Penetration
- Keep GFP in or around plant cells using R9 peptide
- Introduce FT in plant cells using R9 peptide--not yet
4.Activation
- Get high quality of RNA
- Amplify genes successfully
- Check the function of FT--not yet
Future Works
We noticed only flowering and florigen in this time but there are many other plant hormones. We made translocation pathway from E.coli into plant cells, so we will be able to introduce plant hormones into plant cells if E.coli can make them. It means we can control plant growth in any stage through genetically engineered E.coli. In the future that is not so far, we will be able to meddle in plants' growth――germinating, elongation, flowering, and fructification. We human will finally accomplish a technology that control plants perfectly.
Moreover, R9 peptide functions not only plant cell. R9 peptide works on animal cell similarly. It means that we found a pathway into any kinds of cells. R9 peptide tag enables us to introduce proteins into any cells, so we will be able to control all living cells using this technology.
Biosafety
We cooperated with KAIT-Japan and the mark on the left indicates Biosafety Level of our parts.
[1][http://www.ncbi.nlm.nih.gov/pubmed/15695452 Microsugar Chang et al.(2005) "Cellular internalization of fluorescent proteins via arginine-rich intracellular delivery peptide in plant cells" Plant Cell Physiol, 46(3), 482–488]
[2][http://www.ncbi.nlm.nih.gov/pubmed/16155177 Paula Teper-Bamnolker and Alon Samach1.(2005) "The flowering integrator FT regulates SEPALLATA3 and FRUITFULL accumulation in Arabidopsis leaves" The Plant Cell, 17, 2661–2675]
[3][http://www.ncbi.nlm.nih.gov/pubmed/16099980 Philip A. Wigge et al.(2005) "Integration of spatial and temporal information during floral induction in Arabidopsis" Science, 309(5737), 1056-1059]
[4][http://www.mdpi.com/1424-8247/3/4/961/htm Sara Trabulo et al.(2010) "Cell-penetrating peptides—mechanisms of cellular uptake and generation of delivery systems" Pharmaceuticals, 3, 961-993]
[5][http://www.ncbi.nlm.nih.gov/pubmed/15147914 Unnamalai N, Kang BG, Lee.(2004) "Cationic oligopeptide-mediated delivery of dsRNA for post-transcriptional gene silencing in plant cells" FEBS Lett 21, 566(1-3), 307-10]
[6][http://www.ncbi.nlm.nih.gov/pubmed/22683878 Tracy Palmer and Ben C. Berks.(2012) "The twin-arginine translocation (Tat) protein export pathway" Nat Rev Microbiol, 10(7), 483-96]
[7][http://www.ncbi.nlm.nih.gov/pubmed/14966662 Choi JH, Lee SY.(2004) "Secretory and extracellular production of recombinant proteins using Escherichia coli" Appl Microbiol Biotechnol, 64(5), 625-35]
[8][http://www.ncbi.nlm.nih.gov/pubmed/9042754 Miksch G, Fiedler E, Dobrowolski P, Friehs K.(1997) "The kil gene of the ColE1 plasmid of Escherichia coli controlled by a growth-phase-dependent promoter mediates the secretion of a heterologous periplasmic protein during the stationary phase" Arch Microbiol, 167(2-3), 143-50]
[9][http://www.ncbi.nlm.nih.gov/pubmed/11854367 Seibel BA, Walsh PJ.(2002) "Trimethylamine oxide accumulation in marine animals: relationship to acylglycerol storage" J Exp Biol, 205(Pt 3), 297-306]
[10][http://www.ncbi.nlm.nih.gov/pubmed/11123687 Thomas JD, Daniel RA, Errington J, Robinson C.(2001) "Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli" Mol Microbiol, 39(1), 47-53]
[11][http://www.ncbi.nlm.nih.gov/pubmed/3139642 Suit JL, Luria SE.(1988) "Expression of the kil gene of the ColE1 plasmid in Escherichia coli Kilr mutants causes release of periplasmic enzymes and of colicin without cell death" J Bacteriol, 170(10), 4963-4966]
[12][http://www.ncbi.nlm.nih.gov/pubmed/14702305 DeLisa MP, Lee P, Palmer T, Georgiou G.(2004) "Phage shock protein PspA of Escherichia coli relieves saturation of protein export via the Tat pathway" J Bacteriol, 186(2), 366-373]
[13][http://www.ncbi.nlm.nih.gov/pubmed/16099979 Araki, T et al.(2005) “FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex” Science 309(5737), 1052–1056]
 "
"