|
|
| Line 25: |
Line 25: |
| | </div> | | </div> |
| | | | |
| - | | + | <div class="box"> |
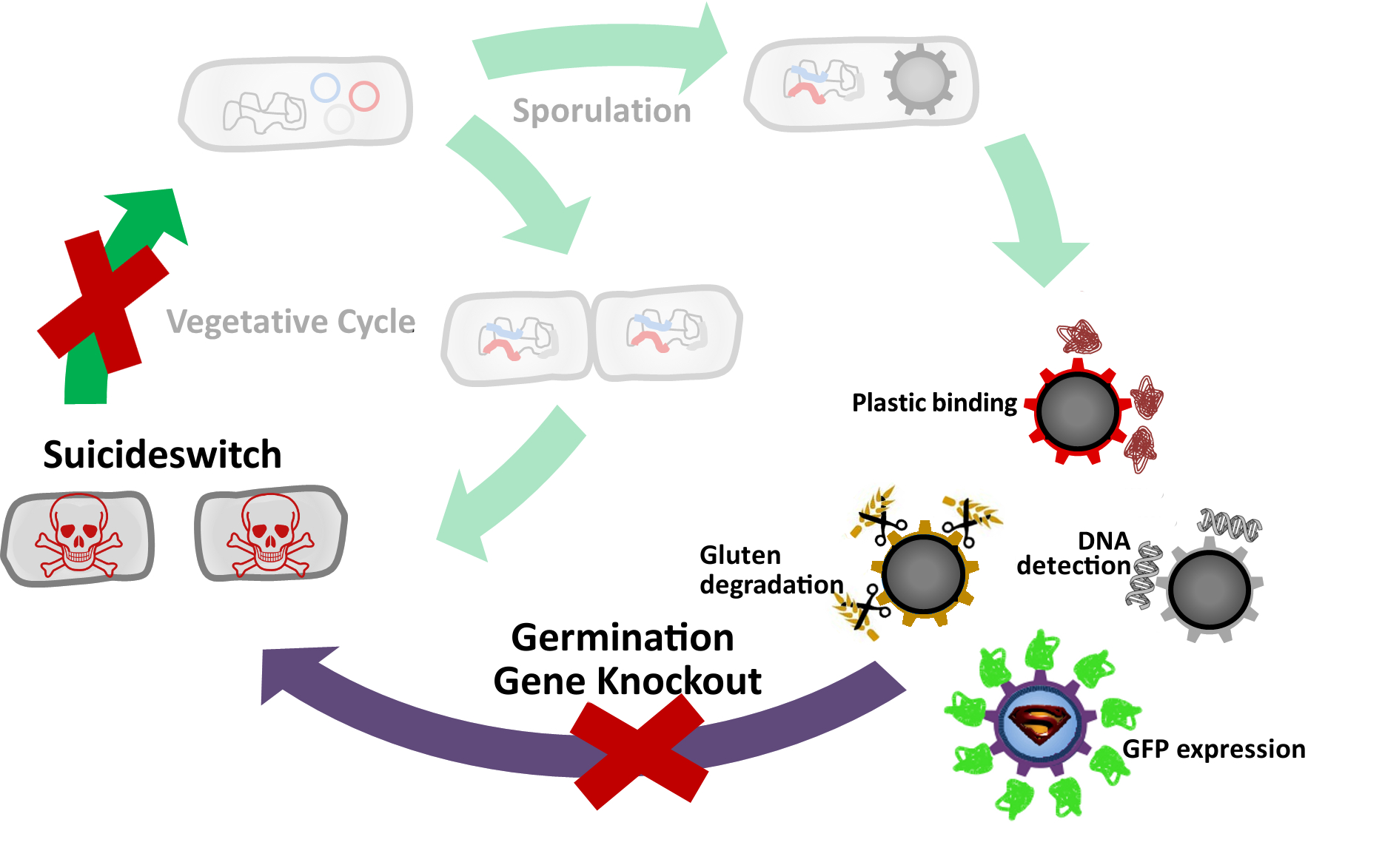
| - | * [[Team:LMU-Munich/Germination_Stop#How do Germination Gene Knockouts Work?|Knock out]] genes that are involved in germination.
| + | |
| - | | + | |
| - | * [[Team:LMU-Munich/Germination_Stop#Suicideswitch|'''Suicide'''switch]]: Toxin production by vegetative cells if germination knockout fails and spores manage to germinate.
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | | + | |
| | =='''Suicide'''switch== | | =='''Suicide'''switch== |
| - | | + | {| "width=100%" style="text-align:center;" style="align:right"| |
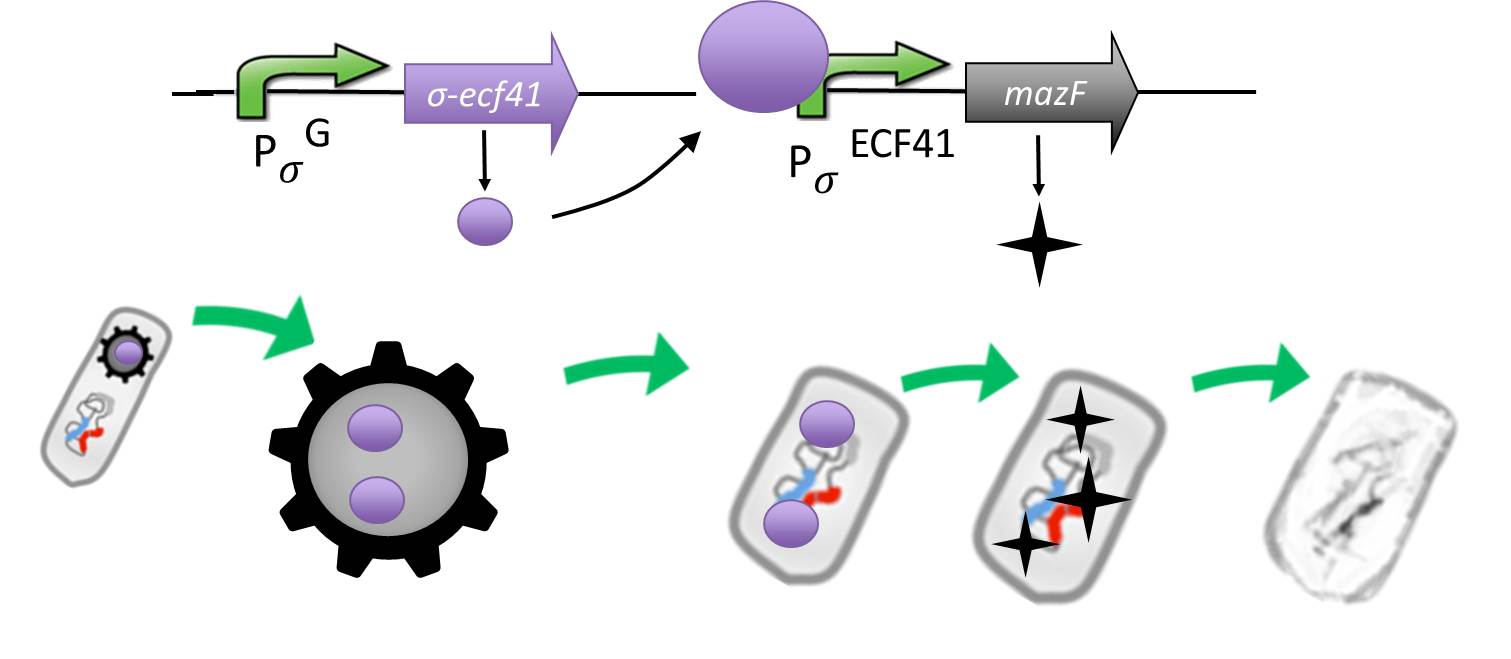
| - | | + | |<p align="justify">If germination knockout fail, we invented the suicide switch. In case spores still germinate, the production of a toxin leads to immediate cell death.</p> |
| - | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | + | |[[File:LMU SuicideSwitch grafik.png|200px|right|link=Team:LMU-Munich/Germination_Stop/SuicideSwitch]] |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | |
| - | {|
| + | |
| - | |[[File:LMU SuicideSwitch grafik.png|624px|center]] | + | |
| | |- | | |- |
| - | | style="width: 70%;background-color: #EBFCE4;" |
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Germination_Stop/SuicideSwitch]] |
| - | {| style="color:black;" cellpadding="3" width="95%" cellspacing="0" border="0" align="left" style="text-align:left;"
| + | |
| - | |style="width: 70%;background-color: #EBFCE4;" | | + | |
| - | <font color="#000000"; size="2">Fig. 5: Genetic elements of the '''Suicide'''switch and its expected performance.</font>
| + | |
| - | |}
| + | |
| - | |}
| + | |
| | |} | | |} |
| | + | </div> |
| | | | |
| | | | |
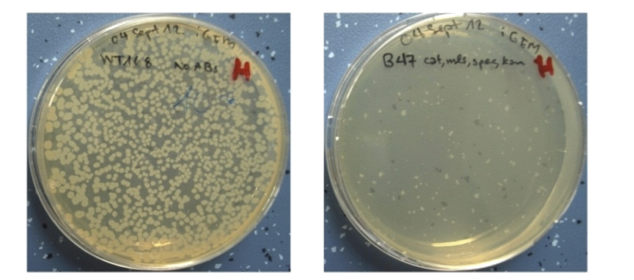
| - | <p align="justify">As a backup plan to make our [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins <b>Sporo</b>beads] even safer, we developed the <b>Suicide</b>switch. In case the spores do germinate, due to degradation or destruction of their outer coats, e.g. by high pressure, the <b>Suicide</b>switch will be turned on.<br></p>
| |
| - | <p align="justify">We take advantage of an alternative and heterologous σ factor and the target promoter. Both are not present in ''B. subtilis'' and therefore not recognized by any other σ factor of ''B. subtilis''.</p>
| |
| - | <p align="justify">The alternative sigma factor [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823043 ecf41<sub>Bli aa1-204</sub>] ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3426412/ Wecke ''et al.'' 2012]), which derives from ''B. licheniformis'' a truncated version constitutively "ON" is synthetically linked to one out of two σ<sup>G</sup>-regulated promoters ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K823048 P<sub>''spoIVB''</sub>] ([http://www.ncbi.nlm.nih.gov/pubmed/15699190 Steil ''et al.'' 2005]) responding strongly or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823042 P<sub>sspK</sub>] ([http://www.ncbi.nlm.nih.gov/pubmed/15699190 Steil, Völker ''et al.'' 2005]) responding weakly). σ<sup>G</sup> is the last σ factor activated in the forespore. Consequently, Ecf41<sub>Bli aa1-204</sub> is produced quite late and only in the forespore. Ecf41<sub>Bli aa1-204</sub> then activates its unique target promoter [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823041 P<sub>''ydfG''</sub>] ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3426412/ Wecke, Mascher 2012]). This promoter is fused to [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823044 MazF] ([http://jcs.biologists.org/content/118/19/4327.abstract Engelberg-Kulka, Amitai 2005]). Activation of P<sub>''ydfG''</sub> therefore leads to the expression of MazF, a bacterial toxin from ''E.coli'' degrading mRNA. <br></p>
| |
| - | <p align="justify">The resulting time course of expression is as follows: Sporulation of the [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins <b>Sporo</b>beads] leads to Ecf41<sub>Bli aa1-204</sub> production in the forespore at the end of the differentiation cycle, through the activation of either P<sub>''spoIVB''</sub> or P<sub>sspK</sub>. Subsequently, this would lead to the expression of MazF, due to the activation of P<sub>''ydfG''</sub>. For our already matured [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins <b>Sporo</b>beads], this would not be toxic, as they represent a dormant state that does not require translation any more. But in the case that a [https://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins '''Sporo'''bead] escapes the '''Germination'''STOP based on the gene knockouts described above, a germinating cell will already contain or soon start producing MazF and therefore kill the vegetative cells.<br></p>
| |
| - | <p align="justify">
| |
| - | See what [https://2012.igem.org/Team:LMU-Munich/Data/Suicideswitch Data] we got when measuring a module with ''luxABCDE'' instead of MazF.
| |
| - | </p>
| |
| - | <p align="justify">
| |
| - | We are planning to model this system, but still need more quantitative, time-resolved data for it. We hope to get this data after finishing the cloning of P<sub>''sspK''</sub> and MazF into ''B. subtilis''.</p>
| |
| | | | |
| | [[File:NEXT.png|right|80px|link=Team:LMU-Munich/Application]] | | [[File:NEXT.png|right|80px|link=Team:LMU-Munich/Application]] |







 "
"