Team:UC-Merced/Notebook
From 2012.igem.org
Darealsunny (Talk | contribs) |
|||
| Line 180: | Line 180: | ||
CaCl 2 solution | CaCl 2 solution | ||
| - | + | [[File:cacl2stck.png]] | |
| - | + | ||
| - | + | ||
| - | + | ||
Need 176 mL = 180 mL so V2 = 180 mL | Need 176 mL = 180 mL so V2 = 180 mL | ||
| Line 242: | Line 239: | ||
2) 1KD FO1 FO2 FO3 100kb AD1 AD2 AD 1kD | 2) 1KD FO1 FO2 FO3 100kb AD1 AD2 AD 1kD | ||
| + | [[File:pcrmx1.png]] | ||
| + | [[File:pcrmx2.png]] | ||
| + | [[File:pcrmx3.png]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</font></td> | </font></td> | ||
</tr> | </tr> | ||
| - | |||
<div align="center"> | <div align="center"> | ||
<table height=75% width=100% border=1 bordercolor=white"> | <table height=75% width=100% border=1 bordercolor=white"> | ||
| Line 287: | Line 253: | ||
<td align=center><font color=black><h4>September 29, 2012:</h4> PCR Gel Agarose Images</font></td> | <td align=center><font color=black><h4>September 29, 2012:</h4> PCR Gel Agarose Images</font></td> | ||
<td align=left><font color=black> | <td align=left><font color=black> | ||
| + | PCR Gel Agarose Images | ||
GE Healthcare: | GE Healthcare: | ||
| Line 300: | Line 267: | ||
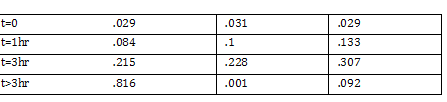
note: step 2 of inoculation to OD540 of 0.375 | note: step 2 of inoculation to OD540 of 0.375 | ||
takes 3.5 hours (estimate) | takes 3.5 hours (estimate) | ||
| - | -placed in -70 degree Celsius freezer | + | -placed in -70 degree Celsius freezer |
| + | |||
| + | |||
| + | </font></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <div align="center"> | ||
| + | <table height=75% width=100% border=1 bordercolor=white"> | ||
| + | |||
| + | <tr> | ||
| + | <td align=center><font color=black><h4>October 1, 2012:</h4> Gibson Ligation</font></td> | ||
| + | <td align=left><font color=black> | ||
| + | |||
| + | Transformation using CaCl2 Trial 2 | ||
| + | |||
| + | 11:40am Inoculated another 400mL of LB+Strephomyocin with 4 mL of FMJ39 | ||
| + | Placed in shaker at 37 degrees Celsius with 250 RPM | ||
| + | |||
| + | 1 @ 590nm wavelength (nanodrop) | ||
| + | |||
| + | [[File:100112.png]] | ||
| + | |||
| + | @ 590nm | ||
| + | |||
| + | Gibson Ligation: | ||
| + | Performed Gibson Ligation using Protocol w/kit | ||
| + | |||
| + | Amount of DNA used: | ||
| + | ADH: 1.4microliter | ||
| + | FO: 1.5 micro liter | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Gibson Master Mix: 10 micro liter | ||
| + | DH20: 7.1 micro liter | ||
| + | Total: 20 micro liter | ||
| + | |||
| + | Gibson fraction gel | ||
| + | Ran Gel using results of Gibson ligation step | ||
| + | |||
| + | -Excised marked band and extracted using gel extraction kit | ||
| + | -namedropped extracted eluted DNA | ||
| + | 14.3 ng/ul, 37.7, 61.1 <-- 3 trial | ||
| + | Average 45ng/ul | ||
| + | |||
| + | Setup PCR on Gibson Ligated Parts | ||
| + | -leave overnight in PCR machine | ||
| + | |||
| + | Standard Parts Assembly | ||
| + | -Set PCR for AD and FO using standard part primers and protocol supplied by Marcos Garcia-Ojeda | ||
| + | |||
| + | -Run and purify on gel | ||
| + | |||
| + | Gel Order (1kb) | ||
| + | 1st piece FO1 FO2 Ladder AD1 AD2 | ||
| + | 2nd piece FO1 FO2 AD1 AD2 | ||
| + | |||
| + | -Collect DNA from gel | ||
| + | |||
| + | Nanodrop results | ||
| + | [[File:1001ND.png]] | ||
| + | |||
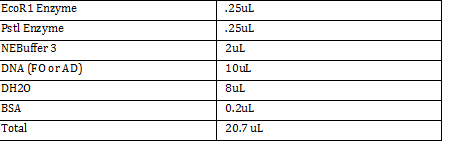
| + | -DNA Digestion of AD and FO | ||
| + | Combine in two tubes: | ||
| + | [[File:rd1001.png]] | ||
| + | |||
| + | |||
| + | Volume in DNA collection tube | ||
| + | |||
| + | FO: 20uL AD: 50uL | ||
| + | |||
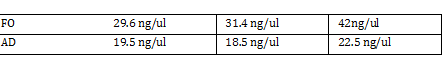
| + | Total DNA taken for restriction enzyme | ||
| + | FO: (10uL)(34.3ng/uL) = 343 ng | ||
| + | |||
| + | AD: (10uL)(20.16ng/ul) = 201.6 ng | ||
| + | |||
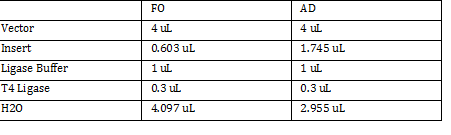
| + | Ligation Step: | ||
| + | |||
| + | 34.3ug/20.7uL = 10ng/x | ||
| + | |||
| + | X = 207uL/34.3 | ||
| + | |||
| + | FO = 0.603 uL | ||
| + | |||
| + | 201.6ug/20.7uL = 17g/x | ||
| + | AD = 1.745 uL | ||
| + | |||
| + | a. RxN Volumes | ||
| + | |||
| + | [[File:v1001.png]] | ||
| + | |||
| + | </font></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <div align="center"> | ||
| + | <table height=75% width=100% border=1 bordercolor=white"> | ||
| + | |||
| + | <tr> | ||
| + | <td align=center><font color=black><h4>October 2, 2012:</h4> Transformation</font></td> | ||
| + | <td align=left><font color=black> | ||
| + | Gel for Gibson-Ligated AD-FO DNA | ||
| + | Gel Well Order | ||
| + | |||
| + | 1kB Ladder Gel G2 | ||
| + | (gene) blank (control) | ||
| + | |||
| + | Excised marked band and extracted using Gel Purification Kit | ||
| + | This band should contain the AD FO gene w/Eco R1 and Pstl site | ||
| + | |||
| + | Nanodrop results- 2.7 ng/uL | ||
| + | |||
| + | -PCR for more Gibson Assembly product done by Israel | ||
| + | |||
| + | G1 is DNA G2 is H2O | ||
| + | Used 1 uL of DNA from 10/1 @45ng/uL | ||
| + | |||
| + | Transformation | ||
| + | -Briefly centrifuged the ligation tubes | ||
| + | -place the tubes on ice | ||
| + | -thaw the competent cells on ice | ||
| + | -add about 50 uL of cells into 4 tubes | ||
| + | -one for FO | ||
| + | -one for AD | ||
| + | -one for straight plasmid | ||
| + | -one for negative control | ||
| + | -Add 5 uL of ligation rxn into corresponding vial | ||
| + | -Tap to mix | ||
| + | -Incubate tubes on ice for 30 minutes | ||
| + | -Incubate tubes @ 47 degrees for 30s | ||
| + | -place tubes back on ice | ||
| + | -Incubate the tubes @ 37 degree Celsius, 300 rpm for 1 hr | ||
| + | -Spread 20-200uL on plates over night | ||
| + | -The on remain cells can be stored @ 4 degree Celsius | ||
| + | |||
| + | |||
| + | |||
</font></td> | </font></td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
Revision as of 02:26, 4 October 2012
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Notebook | Safety | Background | Attributions |
|---|
September 19, 2012:DNA Extraction |
Data: 215 Cells/5 Squares = 43 cells/square x = 43*100*4*10^(-3) x = 43 cells/square (43/.004 mm^3) 1000100 = 1.075*10^9 cells/mL (2.5*10^(7)) cells / (1.075*10^9 cells) = .023mL of bacteria Then start the extraction using the mini-prep kit (insert link to mini prep kit protocol ) Turn on the thermomixer when the buffers are being added -Temp at 56 degree C for 10 minutes -Takes around 5 minutes to heat up Use filter tips since we’re working with DNA As a funny mess up, we ended up throwing away the glass slip for the hemocytometer!!! |
| September 26, 2012: Glycerol stocks + Agar+ Nanodrop |
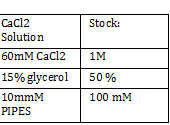
Nanodrop -add small drop of DiH2O 3 times -dab with kim wipe do not wipe -dab both top and bottom bend -log onto ND1000 program and select nucleic acid -add DIH2O with micropipette and close top 2x - add 1 micro liter of DNA to nanodrop, close and measure -save and print Results: 24.1 nanogram/microliter 260/280 = 1.82 260/230 = .49 Remember to clean machine with DI H2O after use! 1 M CaCl2 Protocol -calculate morality of 1M CaCl2 Mass = M*Vol*MW Mass = 14.702 grams per 100 mL of H2O Add 60 L H2O to beaker Add 14.702g CaCl2 to beaker and stir Add back to column and fill up to 100mL Use 150 mL filter until PIPES protocol Weight out 2.307 g of pipes solid Required solution must be at 7pH so add NaOH and measure pH using pH stripes or indicator Once at 7 pH add dH2O until 100mL Use 150 mL filter unit |
September 27, 2012:Antibiotic plates |
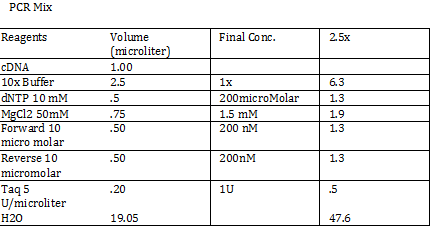
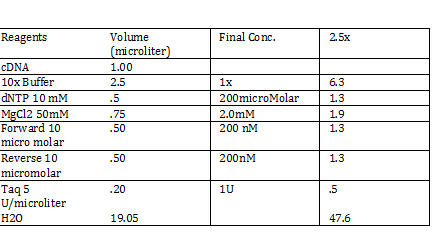
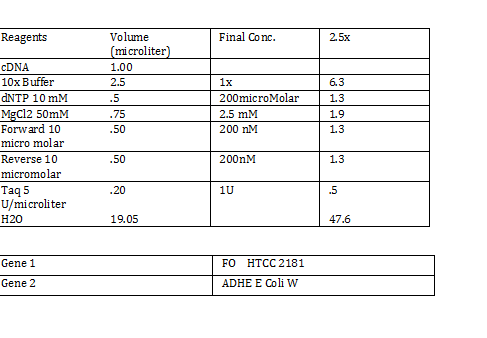
Calculations (Note: Dilution formula was used) Chloramphenicol: (6mg/mL)x=(.025mg/mL)(250mL) x = 1mL Kanamycin: (25 mg/mL)x = (0.05 mg/mL)(250mL) x = .5mL Streptomycin: (20mg/mL)x = (0.02 mg/mL)(250mL) x = .25 mL Antibiotic Plates (Con’t): Data 1. Chloramphencial: Recommended = 25microgram/mL, stock = 6 mg/mL 2. Kanamycin: Recommended = 50 microgram/mL, stock = 25 mg/mL 3. Strephtomycin: Recommended = 20 microgram/mL, stock = 20 mg/mL Note: Color coded the plates: Chloramphenciol (Green) Kanamycin (Orange) Strep/Chloram (Green Black) Bacterial Incubation with Antibiotics E coli W – no antibiotics FMJ 39- strepromycin (25microgram/mL) JW 228- Kanamycin (25 microgram/mL) Bba_K27300- 1000x Amp To make 1000x Amp (25mg/mL)V = (25microgram/mL)(5mL) V = 5 microliter Incubate bacteria in 5mL of LB with antibiotics Glycerol stocks of bacteria Note: Remember to use Aseptic Technique! -Mix 700 micro liter log phase culture with 300 microliter 50 % glycerol -Vortex -Store into cryotube -place in -80 degree C for storage Nanodrop for HTCC 2181 -Same procedure as the one above Results: -260/280: 1.78 -260/230: .17 -2 ng/microliter 260/280: 1.98 260/230: .13 1.4 ng/microliter 260/280: 2.53 260/230: .13 2.9 ng/microliter TAE buffer 1 x for electrophoresis Calculations C1V1 = C2V2 50V1 = (1x)(1000mL) V1 = 20mL of 50x Primer Stocks [100 micromolar primered stocks] -AD BB Pfx Add 633 microliter DH20 -AD GSA Bt Add 1213 micro lite rDH2O -FO GSA Tp Add 1314 microliter DH2O FO BB Sfx Add 615 microliter DH2O Diluted to 10 micromolar working volumes Incubated FMJ39 and JW 1228 for competent because not sure which strain will be used |
 "
"