Team:Calgary/Project/HumanPractices/Killswitch/Regulation
From 2012.igem.org
| Line 57: | Line 57: | ||
<p>Even though this system is a relatively good way to regulate our kill genes, there is some limitation to this system. The main problem why this circuit will not work for our system is because when tailing pond water is added to the bioreactor there is fairly high concentration of Mn2+ in the contaminated water ~48mg/L or ~170µM/L. This concentration is 17 times the amount of Mn2+ needed to trigger the system (10 µM) therefore an additional chemical such as EDTA (a chelator) will have to be added to lower the manganese levels in the bioreactor. This however brings up another situation since EDTA is fairly expensive and will have to be constantly supplied to the bioreactor. Although this system may not be feasible for our system, this regulator system may be used in another pathway.</p> | <p>Even though this system is a relatively good way to regulate our kill genes, there is some limitation to this system. The main problem why this circuit will not work for our system is because when tailing pond water is added to the bioreactor there is fairly high concentration of Mn2+ in the contaminated water ~48mg/L or ~170µM/L. This concentration is 17 times the amount of Mn2+ needed to trigger the system (10 µM) therefore an additional chemical such as EDTA (a chelator) will have to be added to lower the manganese levels in the bioreactor. This however brings up another situation since EDTA is fairly expensive and will have to be constantly supplied to the bioreactor. Although this system may not be feasible for our system, this regulator system may be used in another pathway.</p> | ||
| - | |||
| - | |||
| - | |||
| - | < | + | <h1>Glucose repressible system</h1> |
| - | <p> | + | <h2>Purpose</h2> |
| + | <p>The rhamnose inducible promoter is suitable for controlling the kill switch in the the bioreactor since the promoter is tightly repressed with glucose. We aim to supplement the bioreactor with low levels of glucose to limit expression of the kill genes in the bioreactor. Escape of bacteria into the tailings ponds will cause expression of the kill genes since glucose levels in the surrounding environment would be insufficient for deactivate the promoter. We selected this system for two reasons: firstly, the promoter exhibits tight repression with glucose and could thus limit inadvertent expression of the kill genes; and secondly, glucose is a common monosacharide which is economically feasible to continuously supplement into the bioreactor. | ||
| + | </p> | ||
| - | < | + | <h2>Application of the rhamnose promoter</h2> |
| - | < | + | <p>The rhamnose promoter (pRha) is responsible for regulating six genes related to rhamnose metabolism and contains a separate promoter on its leading and reverse strands (see Figure one). RhaR and RhaS serve to regulate expression of the rhamnose metabolism genes rhaB, rhaA, and rhaD on the opposite side of the promoter. The RhaR transcription factor is activated by L-rhamnose to up-regulate expression rhaSR operon. In turn, the resulting RhaS activates the rhaBAD operon to generate the rhamnose metabolism genes (Egan & Schleif, 1993).</p> |
| - | <p> | + | <p>As a kill switch regulator, our team has harnessed global catabolite repression of the rhamnose promoter. Expression of the rhaBAD operon with RhaS requires the binding of catabolite receptor protein (CRP) cAMP complex to the promoter. When glucose is present, cAMP level are low, such that CRP is unable to activate the promoter (Egan & Schleif, 1993). With this mechanism, our team set out to control our kill genes with glucose with the following rhamnose promoter construct:</p> |
| - | <p> | + | <p>(Draw the final kill circuit)</p> |
| - | <p> | + | <p>Our kill system is different from the native rhamnose system with the rhaR and rhaS control genes. We have constituitively expressed rhaS to overcome dependency on rhamnose to cause activation of the kill switch. While RhaS is continuously present, global catabolite repression prevents its activation the kill genes in the bioreactor; in the outside environment, glucose levels are lower such that RhaS is able to activate the kill genes.</p> |
| - | |||
<h2>Engineering pRha into a kill system</h2> | <h2>Engineering pRha into a kill system</h2> | ||
| - | <p> | + | <p>Components and characterization</p> |
| - | <p> | + | <p>Our team had pRha promoter commercially synthesized as per the sequence given by Jeske and Altenbuchner (2010). The rhaS and rhaR genes were amplified via PCR from Top Ten E. coli using Kapa Hi-Fi polymerase. </p> |
| - | <p> | + | <p>Given the control gene modifications which we have engineered into our system to optimize it for the tailings ponds, we are working to determine whether glucose repression of our modified system match patterns shown by Giacalone et al. (2006) and Jeske and Altenbuchner (2010). To this end, we have constructed the rhamnose promoter with GFP and are finalizing construction with constituitively expressed RhaS, so that we can characterize (LINK) this promoter and test it with our kill genes.</p> |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<h1>The Moco Riboswitch - In progess</h1> | <h1>The Moco Riboswitch - In progess</h1> | ||
<h2>Background</h2> | <h2>Background</h2> | ||
<p> | <p> | ||
| - | |||
| - | |||
The molybdenum cofactor (moco) riboswitch is an RNA element which responds to the presence of the metabolite molybdenum cofactor. This RNA element is located in the E.coli genome just upstream of the moaABCDE operon, which contain the important moco synthesis genes. Moco is an important cofactor in many different enzymes ranging from to this. The moco riboswitch has 2 regions: an aptamer domain and the expression platform. When moco is present in the cell it will bind to the aptamer region in the riboswitch which will cause an allosteric change. This allosteric change affects the expression platform by physically hiding the ribosome binding site thus preventing translation from occurring and hence adding a translational level of gene expression regulation. Therefore, the moco riboswitch, when activated by moco, inhibits gene expression. | The molybdenum cofactor (moco) riboswitch is an RNA element which responds to the presence of the metabolite molybdenum cofactor. This RNA element is located in the E.coli genome just upstream of the moaABCDE operon, which contain the important moco synthesis genes. Moco is an important cofactor in many different enzymes ranging from to this. The moco riboswitch has 2 regions: an aptamer domain and the expression platform. When moco is present in the cell it will bind to the aptamer region in the riboswitch which will cause an allosteric change. This allosteric change affects the expression platform by physically hiding the ribosome binding site thus preventing translation from occurring and hence adding a translational level of gene expression regulation. Therefore, the moco riboswitch, when activated by moco, inhibits gene expression. | ||
Revision as of 04:28, 3 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Regulation/Expression Platform

Magnesium repressible system
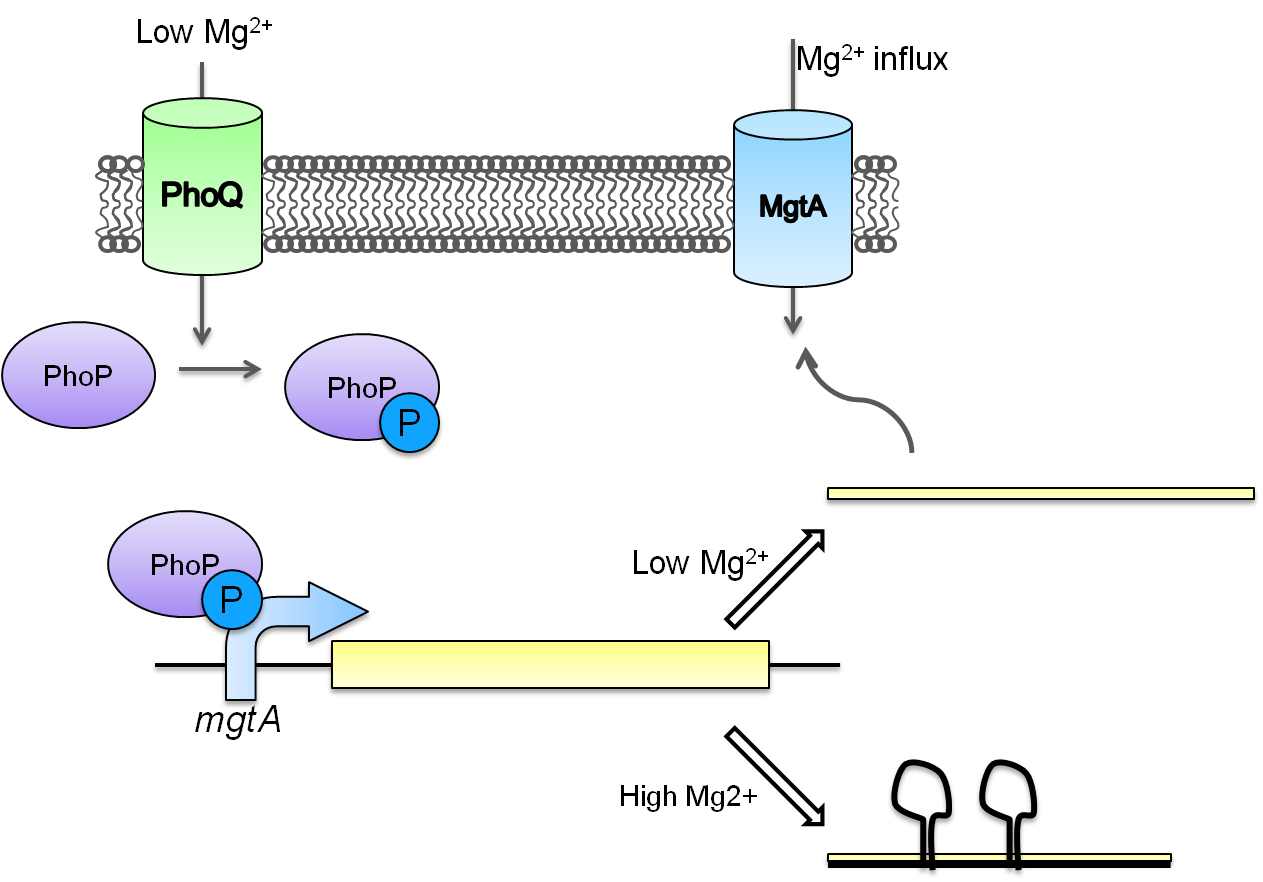
This system is repressed in the presence of magnesium. This system has two control components – a promoter and a riboswitch. Normally the magnesium promoter (mgtA promoter) and the magnesium riboswitch (mgtArb) are activated if there is a deficiency of magnesium in the cell. The lack of magnesium activates other genes in E. coli to allow influx of magnesium into the cell. There are two proteins in the cascade that activate the system namely PhoP and PhoQ. PhoQ is the trans-membrane protein which gets activated in the absence of magnesium and phosphorylates PhoP. PhoP in turn binds to the mgtA promoter and transcribes genes downstream.
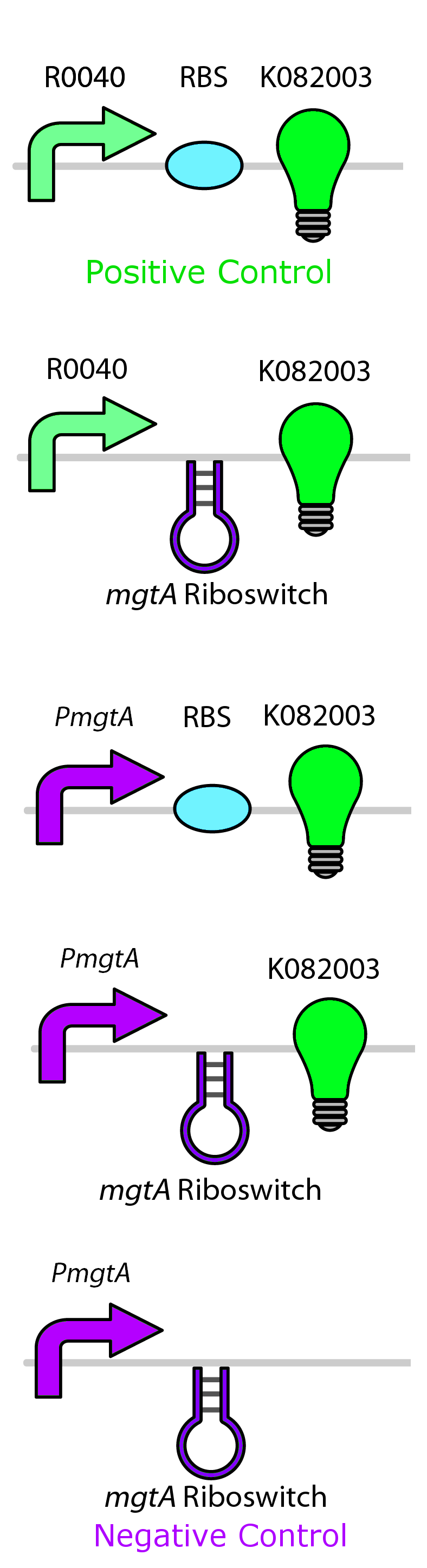
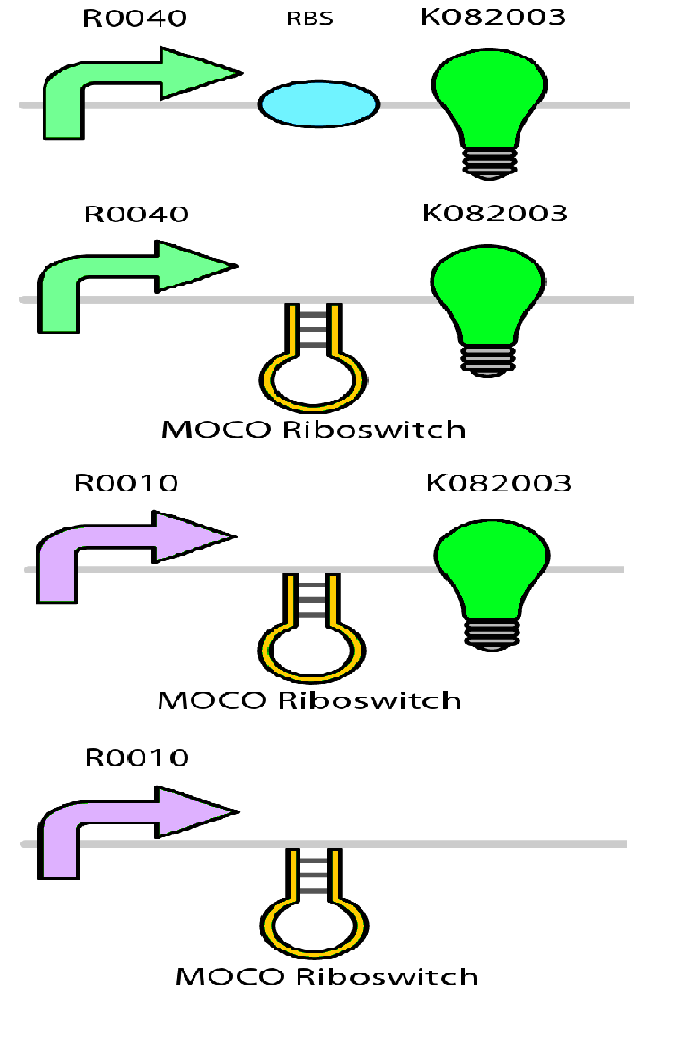
Test circuits for the magnesium system
To test the magnesium regulatory elements we built each of the elements with a reporter gene. We chose Bba_K082003 which is GFP with an LVA tag as our choice of reporter. We did not choose BBa_E0040, the stable GFP, because we wanted a real time indication of the system's control. Stable GFP has a half life of 8 hours and would still fluoresce when the system is shut off.
We build these circuits to test the control elements of the system, namely the mgtA promoter and the mgtA riboswitch.
Characterization of these circuits
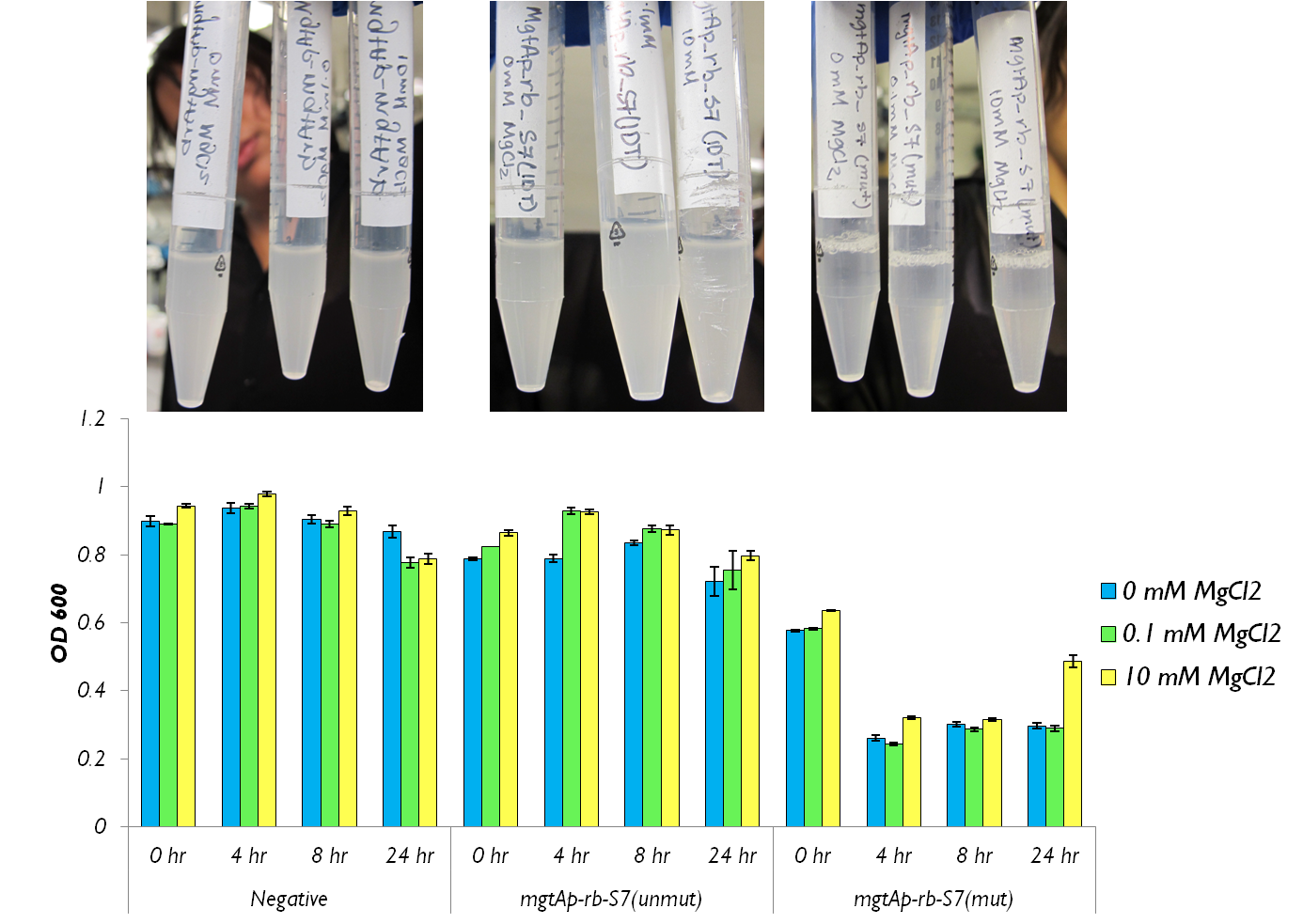
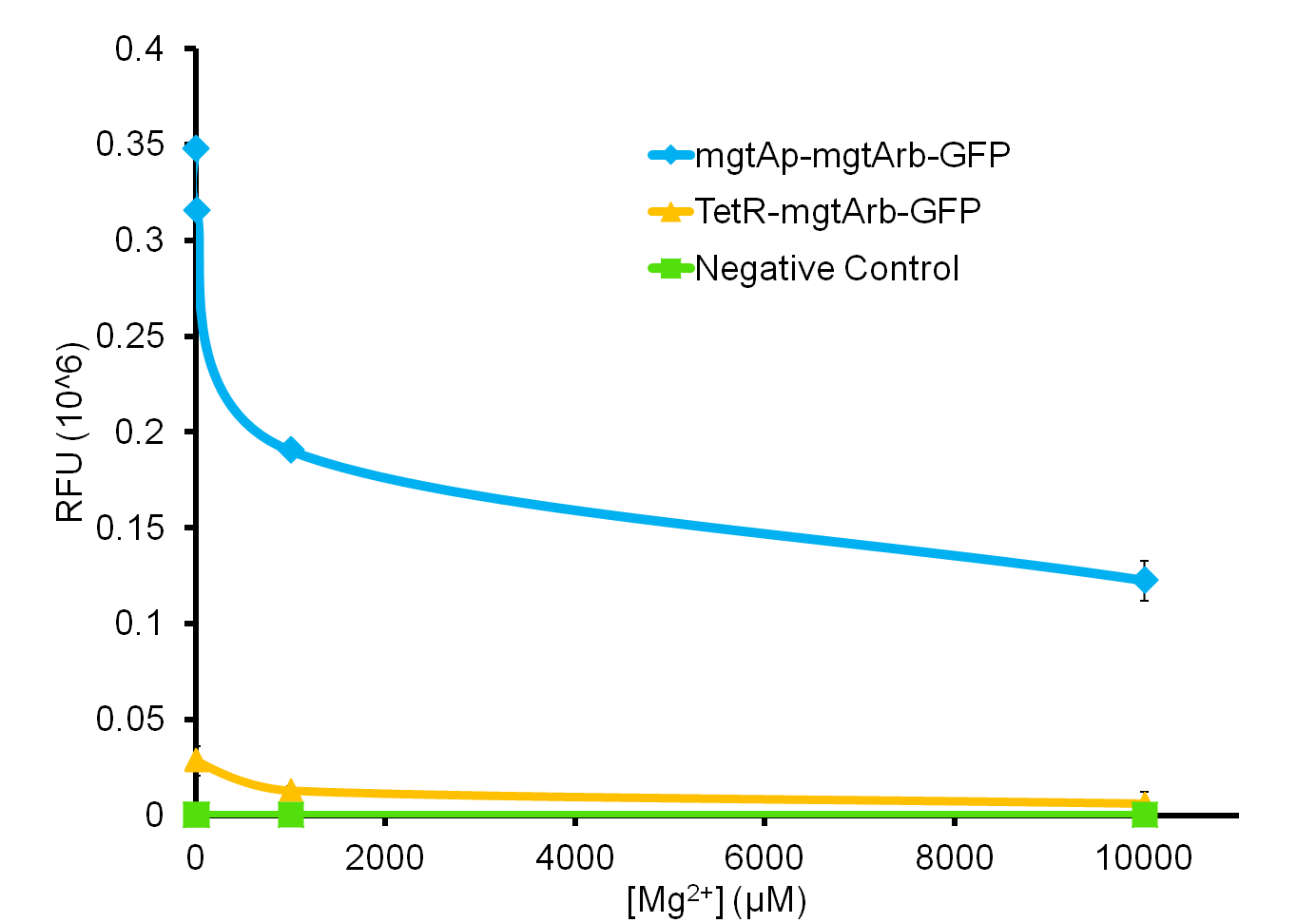
We tested the aforementioned circuits in different concentrations of magnesium. For detailed protocol see INSERT LINK HERE. The values were normalized to the negative control which is the magnesium promoter and riboswitch alone.
There is a much larger decrease in the GFP output when the mgtA promoter and riboswitch are working together compared to the mgtA riboswitch alone under the control of TetR promoter. This suggests that having both the promoter and the riboswitch together provides a tighter control over the genes expressed downstream. This also suggests that magnesium riboswitch alone is sufficient in reducing gene expression downstream of a constitutive promoter.
It is important to consider however that the control elements of the system namely PhoP and PhoQ were not present in the circuits tested and therefore there is some GFP expression in even at the inhibitory concentration (10mM MgCl2). We believe that having the regulatory elements would give us better control and get rid of the leakiness.
Although the magnesium system is highly regulated, it is not a suitable system for the purposes of our bioreactor. The tailings are composed of very high concentration of magnesium- upto 30mM(REFERENCE). As can be seen from figure 3, this would inhibit the system. Therefore, if our bacteria escapes into the tailings, the kill genes would not be activated and the bacteria would be able to survive.
In contrast, it is important to note that this system adds important regulatory elements to the registry such as an inducible promoter and a riboswitch which can be used by other teams to control both killswitches as well as other regulatory pathways which do not pertain using tailings.
We also wanted to test the magnesium system with our kill genes. The micrococcal nuclease that arrived from IDT had 1bp mutation which changed a isoleucine residue into a lysine. Therefore we had to mutate it. To test the circuits we measured both CFU and OD 600.
Manganese regulation
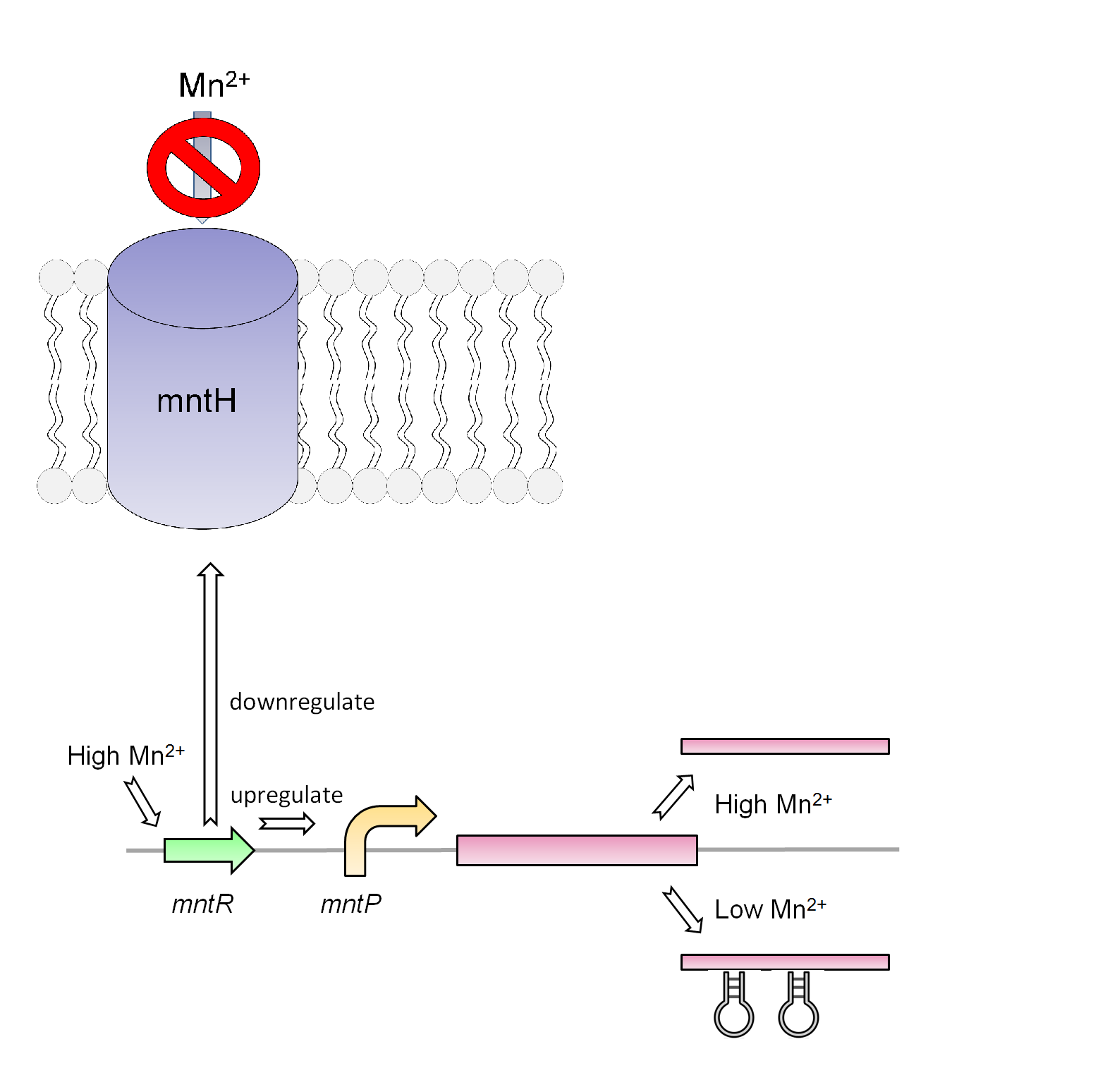
Manganese is found to be an essential nutrient in most bacteria and larger organisms. Due to the properties of manganese this metal can work as a catalyst for many chemical reactions in the body of organisms as well as playing a key role in the structure of macromolecules. Therefore it is not surprising to believe that the cells will need a way to regulate the amount of manganese in its system. Native to the Escherichia coli K-12 strain is the mntR metalloregulatory protein. This transcriptional regulator is found to observe the level of manganese and responds accordingly. The structure of this protein consists of a metal-binding domain that will respond in the presence of manganese and repress the small RNA gene mntS. Lack of mntS will then repress the manganese ion transporter (mntH) as 42-amino-acid protein forms. When this protein membrane is repress the bacteria cannot gained the needed manganese. It is also found that the mntR positively regulates the mntP a putative efflux pump that regulates the intracellular level of manganese.
Our manganese system
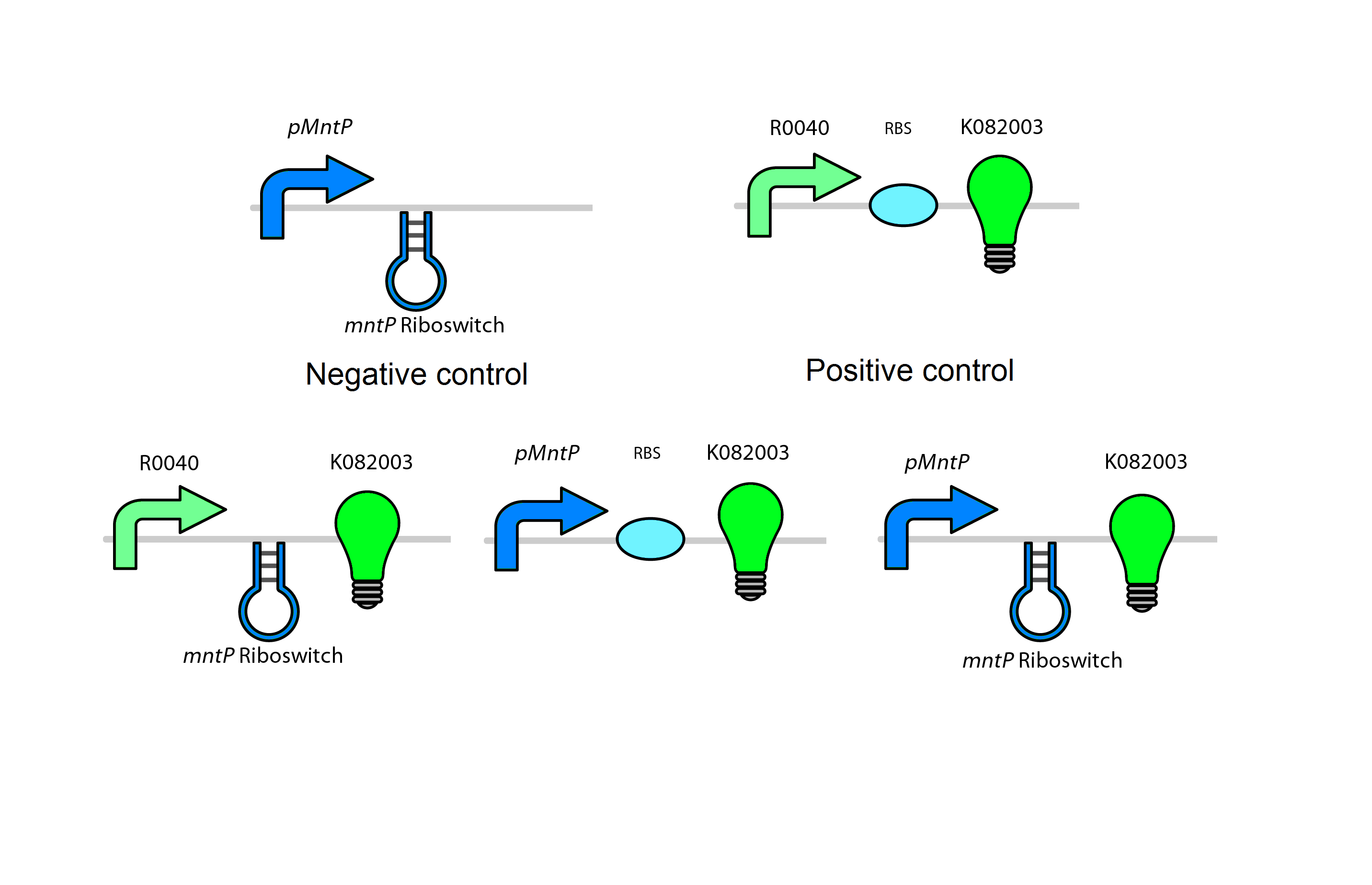
For our system we use the mntR, mntP promoter and mntP riboswitch. As shown in figure _ our system in the present of high Mn2+ will activate the mntR which upregulates the mntP promoter. The mntP promoter in the presence of manganese then initiates the transcription of mntP riboswitch. Likewise in the presence of manganese the mntP riboswitch will express the genes downstream of it or in our case the kill genes S7 and CViAII. In order to control this system in our bioreactor there will have to be low concentrations of Mn2+ in the structure, however when the exposed to the environment or tailing ponds there has to be fairly high concentrations of Mn2+.
Test circuits for the manganese system
The manganese will use a GFP LVA tag. The following are the control circuits built in order to characterise the MntA promoter and the MntA riboswitch.
Future Use
Even though this system is a relatively good way to regulate our kill genes, there is some limitation to this system. The main problem why this circuit will not work for our system is because when tailing pond water is added to the bioreactor there is fairly high concentration of Mn2+ in the contaminated water ~48mg/L or ~170µM/L. This concentration is 17 times the amount of Mn2+ needed to trigger the system (10 µM) therefore an additional chemical such as EDTA (a chelator) will have to be added to lower the manganese levels in the bioreactor. This however brings up another situation since EDTA is fairly expensive and will have to be constantly supplied to the bioreactor. Although this system may not be feasible for our system, this regulator system may be used in another pathway.
Glucose repressible system
Purpose
The rhamnose inducible promoter is suitable for controlling the kill switch in the the bioreactor since the promoter is tightly repressed with glucose. We aim to supplement the bioreactor with low levels of glucose to limit expression of the kill genes in the bioreactor. Escape of bacteria into the tailings ponds will cause expression of the kill genes since glucose levels in the surrounding environment would be insufficient for deactivate the promoter. We selected this system for two reasons: firstly, the promoter exhibits tight repression with glucose and could thus limit inadvertent expression of the kill genes; and secondly, glucose is a common monosacharide which is economically feasible to continuously supplement into the bioreactor.
Application of the rhamnose promoter
The rhamnose promoter (pRha) is responsible for regulating six genes related to rhamnose metabolism and contains a separate promoter on its leading and reverse strands (see Figure one). RhaR and RhaS serve to regulate expression of the rhamnose metabolism genes rhaB, rhaA, and rhaD on the opposite side of the promoter. The RhaR transcription factor is activated by L-rhamnose to up-regulate expression rhaSR operon. In turn, the resulting RhaS activates the rhaBAD operon to generate the rhamnose metabolism genes (Egan & Schleif, 1993).
As a kill switch regulator, our team has harnessed global catabolite repression of the rhamnose promoter. Expression of the rhaBAD operon with RhaS requires the binding of catabolite receptor protein (CRP) cAMP complex to the promoter. When glucose is present, cAMP level are low, such that CRP is unable to activate the promoter (Egan & Schleif, 1993). With this mechanism, our team set out to control our kill genes with glucose with the following rhamnose promoter construct:
(Draw the final kill circuit)
Our kill system is different from the native rhamnose system with the rhaR and rhaS control genes. We have constituitively expressed rhaS to overcome dependency on rhamnose to cause activation of the kill switch. While RhaS is continuously present, global catabolite repression prevents its activation the kill genes in the bioreactor; in the outside environment, glucose levels are lower such that RhaS is able to activate the kill genes.
Engineering pRha into a kill system
Components and characterization
Our team had pRha promoter commercially synthesized as per the sequence given by Jeske and Altenbuchner (2010). The rhaS and rhaR genes were amplified via PCR from Top Ten E. coli using Kapa Hi-Fi polymerase.
Given the control gene modifications which we have engineered into our system to optimize it for the tailings ponds, we are working to determine whether glucose repression of our modified system match patterns shown by Giacalone et al. (2006) and Jeske and Altenbuchner (2010). To this end, we have constructed the rhamnose promoter with GFP and are finalizing construction with constituitively expressed RhaS, so that we can characterize (LINK) this promoter and test it with our kill genes.
The Moco Riboswitch - In progess
Background
The molybdenum cofactor (moco) riboswitch is an RNA element which responds to the presence of the metabolite molybdenum cofactor. This RNA element is located in the E.coli genome just upstream of the moaABCDE operon, which contain the important moco synthesis genes. Moco is an important cofactor in many different enzymes ranging from to this. The moco riboswitch has 2 regions: an aptamer domain and the expression platform. When moco is present in the cell it will bind to the aptamer region in the riboswitch which will cause an allosteric change. This allosteric change affects the expression platform by physically hiding the ribosome binding site thus preventing translation from occurring and hence adding a translational level of gene expression regulation. Therefore, the moco riboswitch, when activated by moco, inhibits gene expression.
Molybdate in TPW
The Killswitch Design
This riboswitch system can be used to design a killswitch mechanism to regulate the expression of our kill genes, CviAII and S7. The basic design of this system includes the kill genes downstream of the riboswitch all of which is under the control of a constitutive promoter. Downstream of this construct is the moa operon constitutively expressed. This entire construct will be present in a low copy plasmid in our bacteria. While moco synthesis is a normal process in the bacteria we wanted to constitutively express the moa operon for two reasons. First, we wanted to up regulate the expression of moco to ensure high enough concentrations of moco capable of inactivating the kill switch when the bacteria are in the bioreactor. Second, the moa operon in the genome is under regulation by the bacteria to maintain equilibrium and therefore we think it might not be reliable in producing the required concentration. The two moa operons will express our metabolite moco, which will activate the riboswitch and represses the kill genes.
Molybdenum, Mo, is a trace element that is required by the bacteria for moco synthesis. Bacteria cannot uptake molybdenum in the elemental form and so uptakes molybdenum in its oxyanion form molybdate, MoO4, using the molybdate transport system. We wanted a system where molybdate is present in the bioreactor permitting moco synthesis (inactivate killswitch) and absent in the tailings pond water (TPW) preventing moco synthesis (activate killswitch). We discovered that molybdenum is indeed present in the TPW.
Concentrations of Mo in the TPW: • [Mo] in the Syncrude and Suncor TPW in 1990: – 0.183 mg/L in the surface region (1m-10m) – 0.045 mg/L in the sludge region (11m-20m)
Molybdate forms when molybdenum is in contact with both water and oxygen. If the bacteria escape they will first enter the surface region where it is possible for the bacteria to encounter molybdate. If molybdate is present in the TPW, then there is a possibility that moco can be synthesized in the escaped bacteria inactivating the kill switch. This dilemma would defeat the purpose of this killswitch mechanism.
The Solution
Upon further literature research we found that molybdate is transported using a molybdate transport system which is coded by the modABCD operon. It has been shown that knocking out mod C, a gene encoding the ATPase of the transporter, enables the transport system dysfunctional and prevents moco synthesis. However, if molybdate is supplemented in high enough concentrations to the bacteria, the bacteria are able to use its sulphate transport system to transport molybdate. This gave rise to the idea of having a mod C knock out strain of bacteria in our system and supplementing it with molybdate allowing moco synthesis (inactivate killswitch) inside the bioreactor. The paper tried 10mM sodium molybdate supplementation in the media whereas that the litreture level states less than 1 uM. Therefore, in the tailings ponds, the concentration is not high enough and molybdate is transported into the cell preventing moco synthesis. This activates the kill switch. Sodium molybdate is expensive and it can be said that on a large bioreactor scale it is impractical to provide. However, there are inexpensive and effective filtration mechanisms available to filter most of it out and reuse it. The filtration also prevents dumping a load of molybdate ions in the TPW.
Characterization
There are two experiments that we want to run with this system. Firstly, the paper did not characterize the concentration of molybdate needed to enter through the sulphate system. So we want to supply e.coli and mod c knockout strain with various concentations of molybdate and measure the optical density and do a cfu assay. Secondly we want to characterize the following system. Flourescence assay for K08 and od and cfyu for kill genes. At the moment the system is still being built.
Biobricks being built:
 "
"