Team:SEU A/Modeling
From 2012.igem.org
(Difference between revisions)
| Line 139: | Line 139: | ||
Select the logistic function used to describe the symmetric S-shaped curve to fit. The expression of the logistic function is:</br> | Select the logistic function used to describe the symmetric S-shaped curve to fit. The expression of the logistic function is:</br> | ||
</html>[[File:Seua_model_exp1_ola1.jpg|90px|center|Seua_model_exp1_ola1]]<html> | </html>[[File:Seua_model_exp1_ola1.jpg|90px|center|Seua_model_exp1_ola1]]<html> | ||
| - | <p | + | <p align="right">(4-1)</p></br> |
Where A,B,C are unknown parameters.</br> | Where A,B,C are unknown parameters.</br> | ||
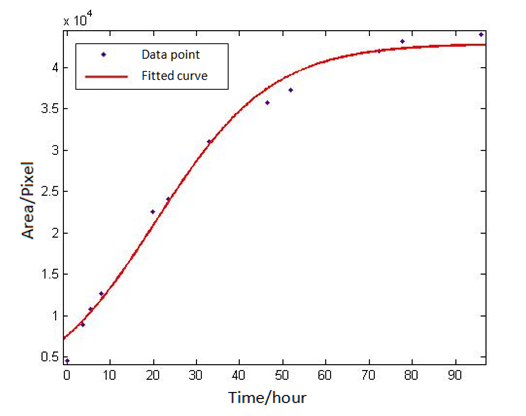
Use ‘cftool’ in Matlab to fit the eight groups of data of the eight plaques, with the default method of least squares. After selecting the appropriate initial value for the three parameters, the results can be obtained quickly. By the fitting of eight groups of data, we found all the right parameters A,B,C to make data points distributing on both sides of the fitted curve and variance is small. If so, it is considered that the eight groups of data conform to the S-shaped curve, and that the image processing above is reliable | Use ‘cftool’ in Matlab to fit the eight groups of data of the eight plaques, with the default method of least squares. After selecting the appropriate initial value for the three parameters, the results can be obtained quickly. By the fitting of eight groups of data, we found all the right parameters A,B,C to make data points distributing on both sides of the fitted curve and variance is small. If so, it is considered that the eight groups of data conform to the S-shaped curve, and that the image processing above is reliable | ||
Revision as of 22:54, 26 September 2012

modeling
The ebb and flow of E. coli in computer..
Index |
1 Analysis of E.coli and Bdellovibrio’s growth in the number of speed by a group of photographs
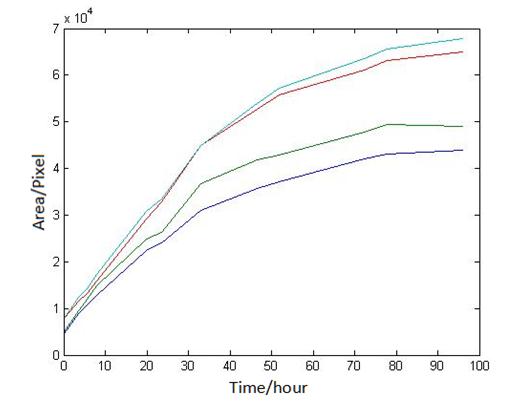
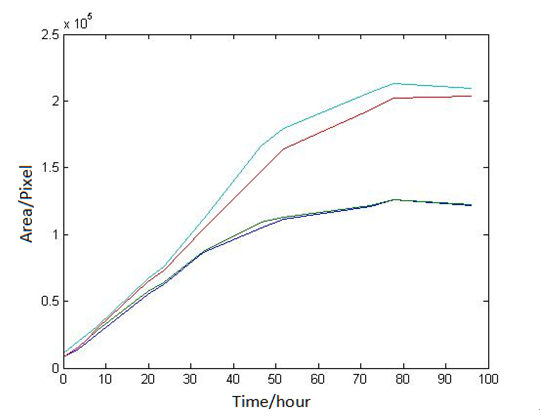
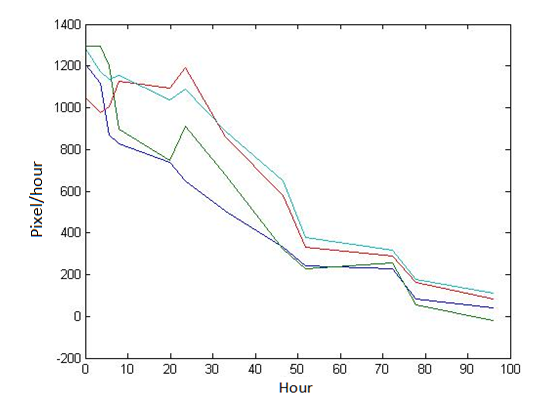
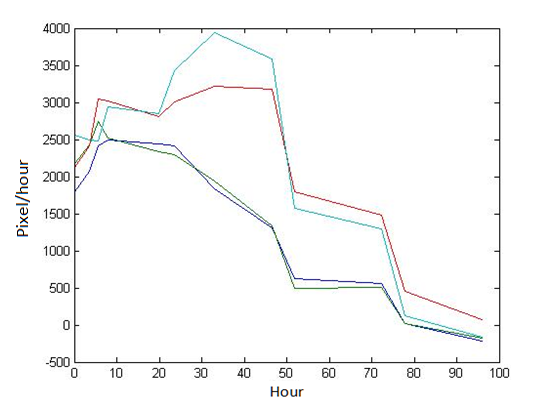
Abstract: Based on the observations at intervals of several days, and the photos of the plaque in the process of E. coli and Bdellovibrio group’s growth in the same dish taken at a high resolution for experimental, we use of the image processing functions of Matlab to simplify the statistics and calculate the relative quantities of the two bacteria at the observation time in the dish. The results also verify whether the experimental results conform to the classical S-shaped curve, and estimate the bacterial growth rate in every moment, in order to analyze the bacterial growth’s features under the conditions of the experiments. Key words: Bacterial growth High resolution photos Image processing S-shaped curve fitting. 1.1 The classical model of the flora growth Under normal circumstances, a growth curve can be obtained with the time as abscissa and the logarithmic of the number of viable cells as ordinate. The curve shows the four periods of the bacterial growth and reproduction: the lag period, the logarithmic period, the stable period, the decline period. The lag period is the transient process the bacteria adapt to the new environment after being inoculated to the medium. In this period, the curve is flat and stable because of slow bacterial multiplication. The number of viable bacteria will soar when it reaches the logarithmic period. The bacteria will grow at stable geometric series, and can last sustainably hours and days (depending on the conditions and bacterial generation). Moreover, bacteria’s form, dyeing and biological activity in this period are all very typical which is sensitive to environmental factors, so it’s the best period to research the bacterial traits. When it reaches the stable period, however, the total number of flora is in the flat stages due to the relative relationship of nutrition and the number of bacteria, but bacterial viability changes greatly. Finally, bacterial growth will get slower in the decline period, and the number of the dead bacteria will apparently increase. In addition, the number of viable cells and the incubation time are in inversely proportional relationship. Mastering the bacterial growth’s law, we can study the method of controlling the growth of pathogenic bacteria, discovering and developing bacteria useful to human. 1.2 Assumptions and simplifications 1. Imagine that plaque is dense, that is to say, it’s always a simply connected area in surface of the two-dimensional dish. 2. Imagine that the plane occupied by the plaque in the culture dish is proportional to the number of bacteria in it. 3. Under the experimental conditions, imagine that E. coli is the only food of Bdellovibrio, and nutrients of E. coli are provided by the medium sufficiently. 4. The occasional small amount of bacteria in the edge of dish is ignored, and dripping or splashing a small amount of bacteria into the location of the error place in the initial state is considered to have no impact on the normal experimental bacteria. 5. After appropriate image processing, the light and its changes’ effects to the photos are ignored. 6. As to the fuzzy edge of the plaque, the area is considered to be the largest and smooth edges simply connected region. 1.3 Assumptions 3.1 Photos’ preprocessing 1. Select photos artificially. Remove the ones whose light effects apparently different from most of the photos, as well as the ones which appear the situation that two plaque overlap and stick together and the overlapping area is large in the later observations, so as not to increase the difficulties of unified image processing. 2. Rename all the photos by unified format. The format is ‘experimental group + days + hours + minutes xxx’, (such as ‘3111111’ meaning the third group at 11:11 on the 11th), which is beneficial to the unified automatic processing. 3. The few overlapping points of two plaques are considered to have no effects on these two, so use Photoshop to change the connections as the background color, in order to processing uniformly. 3.2 Calculation of the plaques’ area 1. After reading the photos, select the appropriate threshold to transform the photos into a binary image, according to the light conditions. 2. Identify all the connected regions, and remove the smaller ones (maybe caused by useless bacteria or lens’ dust). 3. Because of the light and its reflection in every photo, the edges of culture dishes are always lit, which require removing the large connective regions in the edges. 4. Calculate the area of the other connective regions, which is all the plaque area. 5. Use the area above as the relative number of bacteria, and pixel as the unit. Similarly, the number of bacteria growth rate is in unit of pixel/ unit time. And it is the same way if ‘the number of bacteria’, ‘the relative quantities of the bacteria’ and ‘the rate of bacteria growth’ in the paper is not particularly described. 1.4 Results and analysis 4.1 Figure of the changes in the number of E. coli and Bdellovibrio(4-1)
Where A,B,C are unknown parameters. Use ‘cftool’ in Matlab to fit the eight groups of data of the eight plaques, with the default method of least squares. After selecting the appropriate initial value for the three parameters, the results can be obtained quickly. By the fitting of eight groups of data, we found all the right parameters A,B,C to make data points distributing on both sides of the fitted curve and variance is small. If so, it is considered that the eight groups of data conform to the S-shaped curve, and that the image processing above is reliable The fitting results of a particular group are as follows:1 Exp.2
2.1 Summary 2.2 Model 2.3 Assumptions 2.4 Solution 2.5 Evaluation 2.6 ReferencesBiomedical Engineer School, SEU | iGEM 2012
Copyright © Southeast University iGEM 2012 Team A, All rights reserved.
 "
"