Team:LMU-Munich/Spore Coat Proteins
From 2012.igem.org
| Line 42: | Line 42: | ||
{| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
|style="width: 70%;background-color: #EBFCE4;" | | |style="width: 70%;background-color: #EBFCE4;" | | ||
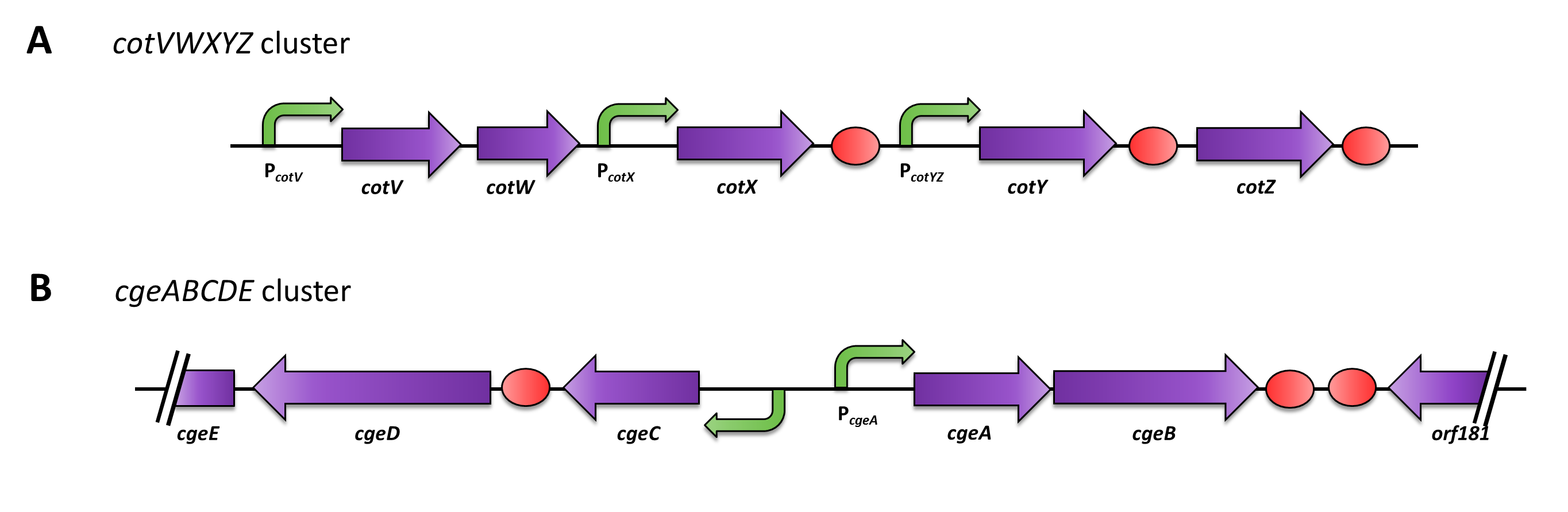
| - | <font color="#000000"; size="2">Gene clusters of ''cotZ'' and ''cgeA''</font> | + | <font color="#000000"; size="2">Fig. 2: Gene clusters of ''cotZ'' and ''cgeA''</font> |
|} | |} | ||
|} | |} | ||
| Line 48: | Line 48: | ||
| - | <p align="justify"> | + | <p align="justify">First, we used [http://partsregistry.org/Part:BBa_K823039 ''gfp''] as a proof of principle and fused it to ''cgeA'' and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823031 ''cotZ'']. This way we would determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation.</p> |
| - | <p align="justify"> | + | <p align="justify">We first constructed the BioBrick for ''cotZ'', ''cgeA'' and ''gfp'' in [http://partsregistry.org/Help:Assembly_standard_25 Freiburg Standard]. ''cotZ''was then fused to its two native promoters, P<sub>''cotV''</sub> and to P<sub>''cotYZ''</sub>, and P<sub>''cgeA''</sub>, which regulates the transcription of ''cgeA''. For ''cgeA'' we only used its native promoter P<sub>''cgeA''</sub> and the stronger one of the two promoters of the ''cotVWXYZ'' cluster, P<sub>''cotYZ''</sub> (for more details see [https://2012.igem.org/Team:LMU-Munich/Data/crustpromoters crust promotor evaluation]. While [http://partsregistry.org/Part:BBa_K823039 ''gfp''] was ligated to the terminator B0014 (see [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0014 Registry]). When these constructs were finished and confirmed by sequencing, we fused them together by applying the Freiburg standard to create constructs, in which gfp is fused C-terminally to ''cotZ'' or ''cgeA'' flanked by one of the three promoters and the terminator. This way we created C-terminal fusion proteins. |
| - | <br>But as we did not know if C- or N-terminal fusion would influence the fusion protein expression, our second aim was to construct N-terminal fusion proteins as well. For this purpose we wanted to fuse the genes for the crust proteins ''cotZ'' and ''cgeA'' to the terminator and ''gfp'' to the three chosen promoters. | + | <br>But as we did not know if C- or N-terminal fusion would influence the fusion protein expression, our second aim was to construct N-terminal fusion proteins as well. For this purpose we wanted to fuse the genes for the crust proteins ''cotZ'' and ''cgeA'' to the terminator and ''gfp'' to the three chosen promoters. Unfortunately, there occured a mutation in the XbaI site during construction of ''gfp'' in Freiburg Standard. Therefore we were not able to finish these constructs. |
</p> | </p> | ||
| Line 66: | Line 66: | ||
{| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
|style="width: 70%;background-color: #EBFCE4;" | | |style="width: 70%;background-color: #EBFCE4;" | | ||
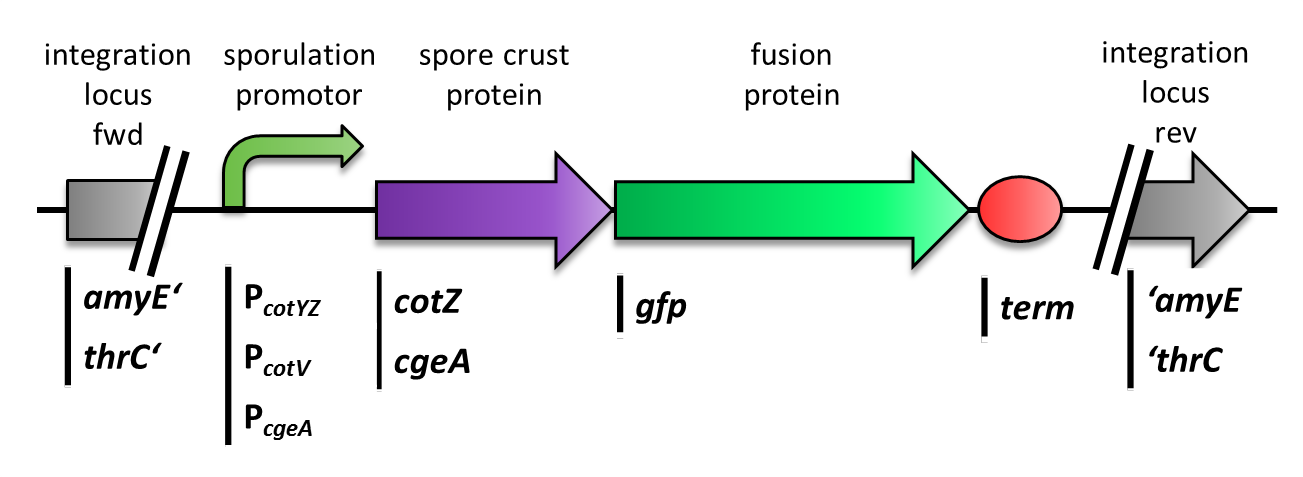
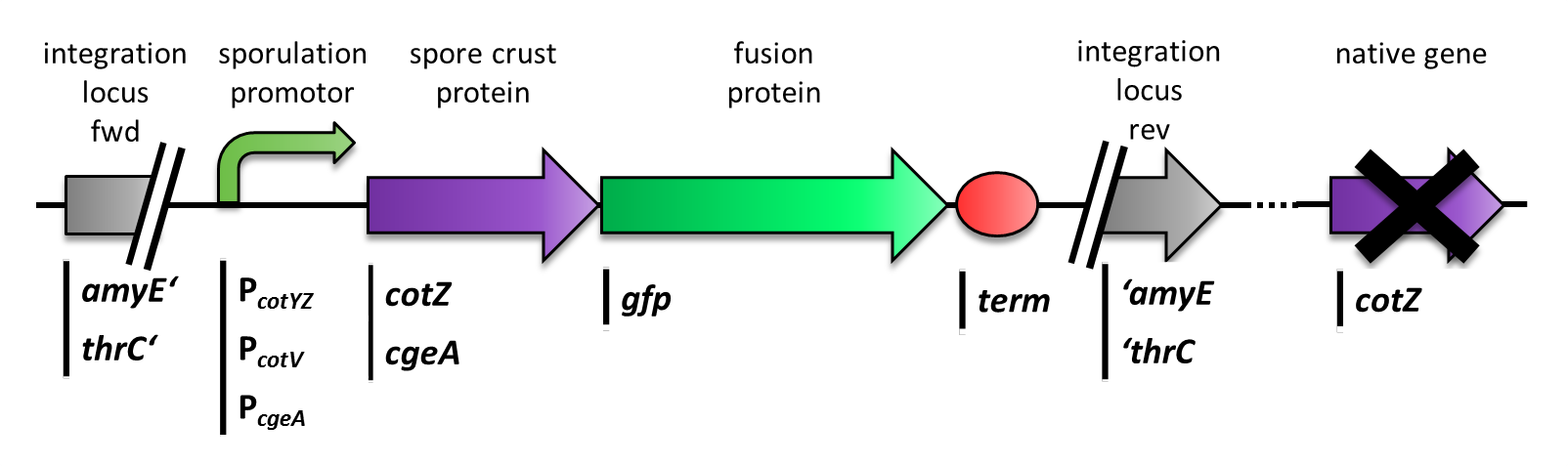
| - | <font color="#000000"; size="2">Section of the genome of ''B. subtilis'' with the various integrated constructs.</font> | + | <font color="#000000"; size="2">Fig. 3: Section of the genome of ''B. subtilis'' with the various integrated constructs.</font> |
|} | |} | ||
|} | |} | ||
| - | <p align="justify"> | + | <p align="justify">Finally we needed to clone our constructs into an empty ''Bacillus'' vector, so that they could get integrated into the genome of ''B. subtilis'' after transformation. Thus we picked the empty vector pSB<sub>BS</sub>1C from our [https://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors '''''Bacillus''B'''io'''B'''rick'''B'''ox], for the ''cotZ'' constructs. This vector integrates into the ''amyE'' locus, which allows to easily check the integration via starch test. In oder to also express both crust protein constructs in one strain, the ''cgeA'' fusion proteins had to be cloned into one of our other empty vectors pSB<sub>BS</sub>4S. Unfortunately for unknown reasons, the cloning of the constructs with ''cgeA'' into this vector has so far not been successful.</p> |
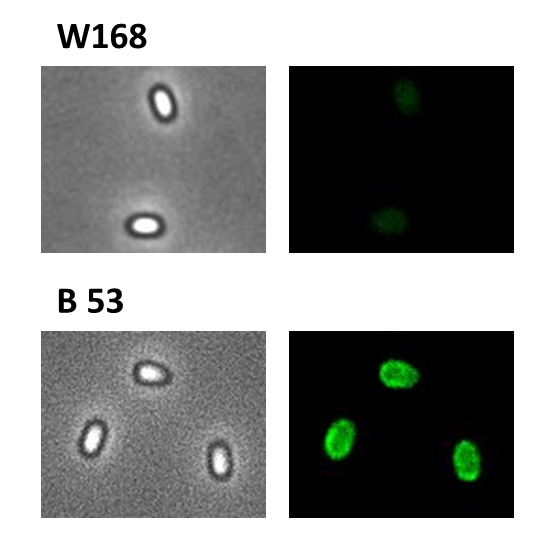
| - | <p align="justify">Finally we | + | <p align="justify">Finally, we started with the most important experiment for our GFP-'''Sporo'''beads, the fluorescence microscopy. Therefore we developed a sporulation protocol(for details see [https://static.igem.org/mediawiki/2012/e/e9/LMU-Munich_2012_Protocol_for_enhancement_of_mature_spore_numbers.pdf Protocol for enhancement of mature spore numbers]), that increases the rates of mature spores in our mutant samples. The cells were fixed on agarose-pads and imaged in bright field and excited in blue wavelength. All '''Sporo'''beads showed green fluorescence on their surface. But B53-'''Sporo'''bead (containing the P<sub>''cotYZ''</sub>-''cotZ''-''gfp''-terminator construct) illusidated the highest fluorescence intensity (see Figure 4). For further experiments, we chose this as it showed the brightest fluorescence.</p> |
| Line 84: | Line 84: | ||
{| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
|style="width: 70%;background-color: #EBFCE4;" | | |style="width: 70%;background-color: #EBFCE4;" | | ||
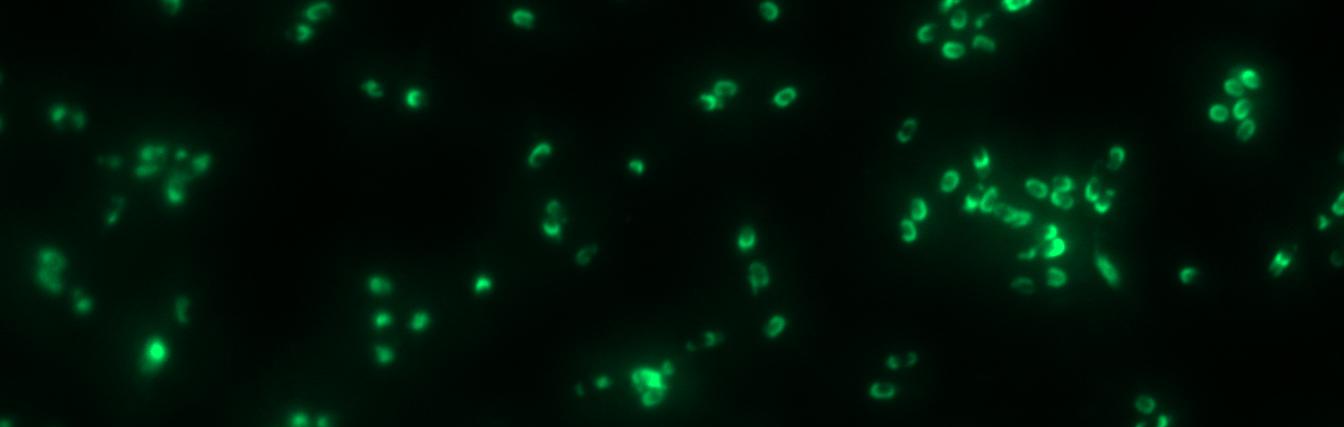
| - | <font color="#000000"; size="2">Fluorescence of wildtype | + | <font color="#000000"; size="2">Fig. 4: Fluorescence of wildtype spores and B53-'''Sporo'''beads (containing the P<sub>''cotYZ''</sub>-''cotZ''-''gfp''-terminator construct)</font> |
|} | |} | ||
|} | |} | ||
| Line 109: | Line 109: | ||
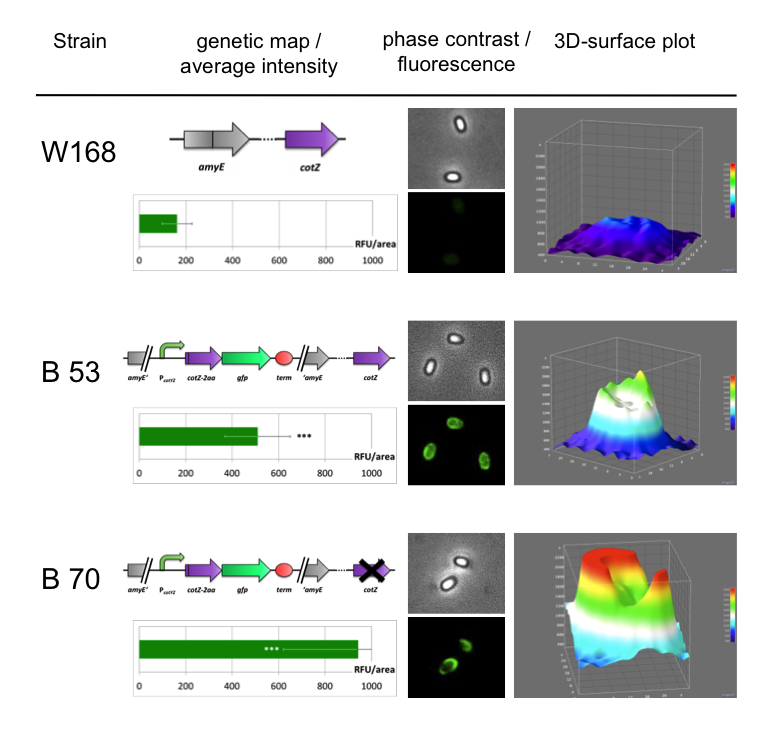
| - | <p align="justify">The '''Sporo'''beads | + | <p align="justify">Because of the low but distinct fluorescence of wildtype spores, we measured and compared the fluorescence intensity of 100 spores per construct. The '''Sporo'''beads were investigated by fluorescence microscopy and analysed. We obtained significant differences between wildtype spores and all our '''Sporo'''beads [see https://2012.igem.org/Team:LMU-Munich/Data/gfp_spore Data]. The intensity bar charts should thereby show the fluorescence difference between wildtype (W168), B53- and B70-'''Sporo'''beads (B53 strain with native ''cotZ'' deletion). To demonstrate the distribution of the fusion proteins we created 3D graphs, which show the fluorescence intensity spread across the spore surface. For analysis we measured the fluorescence intensity of a area of 750px per spore by using ImageJ and evaluated it with the statistical software '''R'''. The following graph shows the results of microscopy and ImageJ analysis.</p> |
Revision as of 21:10, 26 September 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Sporobeads - what protein do you want them to display?
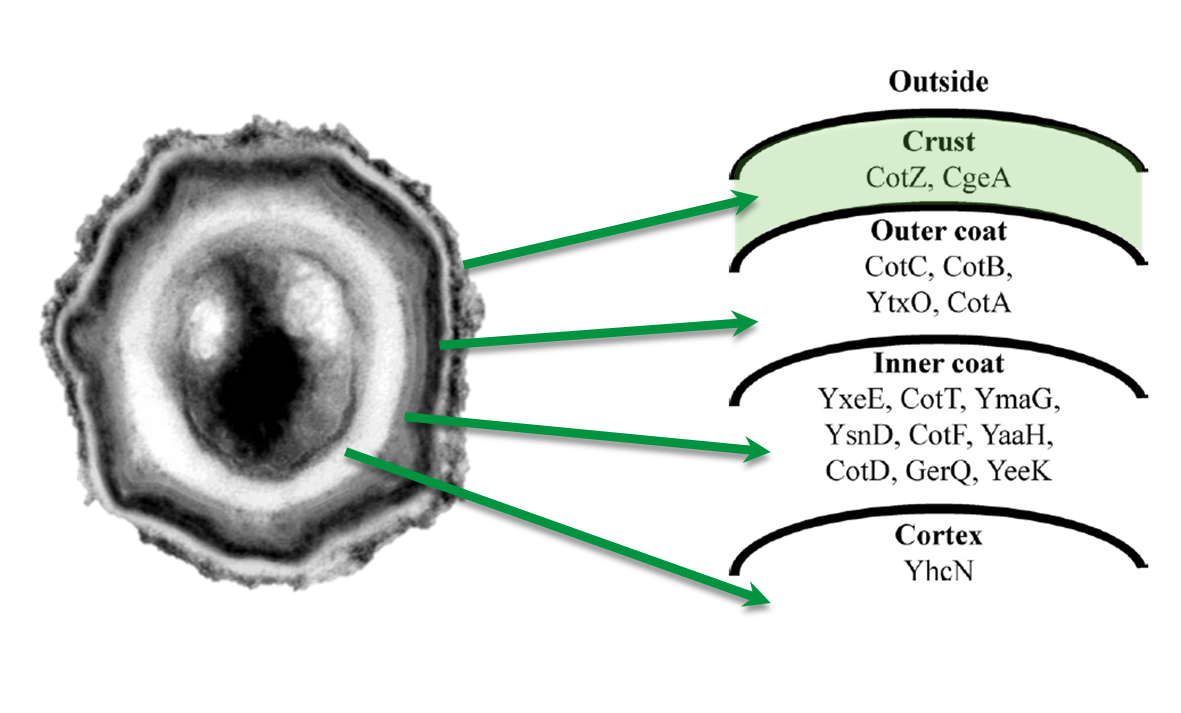
Sporobeads are Bacillus subtilis spores modulated in a way that they can be used as a platform for individual protein display. The aim of this module is to create these spores that display fusion proteins on their surface. There are several different proteins forming the spore coat layers of Bacillus subtilis spores. The outermost layer, the so called spore crust, is composed of two proteins, CotZ and CgeA ([http://www.ncbi.nlm.nih.gov/pubmed?term=imamura%20et%20al.%202011%20spore%20crust Imamura et al., 2011]). This is why we used them to create functional fusion proteins to be expressed on our Sporobeads.
|
The gene cgeA is located in the cgeABCDE cluster and is regulated by its own promoter PcgeA. The cluster cotVWXYZ contains the gene cotZ which is cotranscribed with cotY and regulated by the promoter PcotYZ. Another promoter of this cluster PcotV is responsible for the transcription of the other three genes. Those three promoters were evaluated with the lux reporter genes to get an impression of their strength and their time of activation (see for more details [http://partsregistry.org/Part:BBa_K823025 pSBBs3C-luxABCDE]) so they could be used for expression of spore crust fusion proteins.
|
First, we used [http://partsregistry.org/Part:BBa_K823039 gfp] as a proof of principle and fused it to cgeA and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823031 cotZ]. This way we would determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation.
We first constructed the BioBrick for cotZ, cgeA and gfp in [http://partsregistry.org/Help:Assembly_standard_25 Freiburg Standard]. cotZwas then fused to its two native promoters, PcotV and to PcotYZ, and PcgeA, which regulates the transcription of cgeA. For cgeA we only used its native promoter PcgeA and the stronger one of the two promoters of the cotVWXYZ cluster, PcotYZ (for more details see crust promotor evaluation. While [http://partsregistry.org/Part:BBa_K823039 gfp] was ligated to the terminator B0014 (see [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0014 Registry]). When these constructs were finished and confirmed by sequencing, we fused them together by applying the Freiburg standard to create constructs, in which gfp is fused C-terminally to cotZ or cgeA flanked by one of the three promoters and the terminator. This way we created C-terminal fusion proteins.
But as we did not know if C- or N-terminal fusion would influence the fusion protein expression, our second aim was to construct N-terminal fusion proteins as well. For this purpose we wanted to fuse the genes for the crust proteins cotZ and cgeA to the terminator and gfp to the three chosen promoters. Unfortunately, there occured a mutation in the XbaI site during construction of gfp in Freiburg Standard. Therefore we were not able to finish these constructs.
|
| |
|
Finally we needed to clone our constructs into an empty Bacillus vector, so that they could get integrated into the genome of B. subtilis after transformation. Thus we picked the empty vector pSBBS1C from our BacillusBioBrickBox, for the cotZ constructs. This vector integrates into the amyE locus, which allows to easily check the integration via starch test. In oder to also express both crust protein constructs in one strain, the cgeA fusion proteins had to be cloned into one of our other empty vectors pSBBS4S. Unfortunately for unknown reasons, the cloning of the constructs with cgeA into this vector has so far not been successful.
Finally, we started with the most important experiment for our GFP-Sporobeads, the fluorescence microscopy. Therefore we developed a sporulation protocol(for details see Protocol for enhancement of mature spore numbers), that increases the rates of mature spores in our mutant samples. The cells were fixed on agarose-pads and imaged in bright field and excited in blue wavelength. All Sporobeads showed green fluorescence on their surface. But B53-Sporobead (containing the PcotYZ-cotZ-gfp-terminator construct) illusidated the highest fluorescence intensity (see Figure 4). For further experiments, we chose this as it showed the brightest fluorescence.
|
Since there were still some vegetative cells left after 24 hours of growth in DS-Medium, we wanted to purify the Sporobeads from them, which thereby should be deadened. We chose three different methods for this approach, the treatment with French Press, ultrasound (sonification) or lysozyme. By means of the microscopy results we were able to conclude that lysozyme treatment was the only successful method [link to data]. Additionally, it did not harm the crust fusion proteins as green fluorescence was detectable afterwards [link zu data]. This is why we use this treatment for purifying spores since.</p>
Eventually, clean deletions of the native genes should reveal if there is any difference in fusion protein expression in our Sporobeads. Thus we deleted the native cotZ and cgeA using the cloning method described by [http://www.ncbi.nlm.nih.gov/pubmedterm=New%20Vector%20for%20Efficient%20Allelic%20Replacement%20in%20Naturally%20Nontransformable%2C%20Low-GC-Content%2C%20Gram-Positive%20Bacteria Arnaud et al., 2004].
|
| |
|
Because of the low but distinct fluorescence of wildtype spores, we measured and compared the fluorescence intensity of 100 spores per construct. The Sporobeads were investigated by fluorescence microscopy and analysed. We obtained significant differences between wildtype spores and all our Sporobeads [see https://2012.igem.org/Team:LMU-Munich/Data/gfp_spore Data]. The intensity bar charts should thereby show the fluorescence difference between wildtype (W168), B53- and B70-Sporobeads (B53 strain with native cotZ deletion). To demonstrate the distribution of the fusion proteins we created 3D graphs, which show the fluorescence intensity spread across the spore surface. For analysis we measured the fluorescence intensity of a area of 750px per spore by using ImageJ and evaluated it with the statistical software R. The following graph shows the results of microscopy and ImageJ analysis.
|
| |
|
Applications
There are many possible applications for our Sporobeads as it is possible to modify their outermost protein layer. This way they could be used as beads with special functions. To easily create any kind of Sporobead we designed a Sporovector, where you just have to insert your protein of choice. Since there are so many possible applications we picked three examplary ideas for future Sporobeads:
Kumamolisin-Sporobeads: the solution for carefree enjoyment of everyday meals
World wide one out of [http://www.enriquecastro.net/index.php/term/,9da4ab975b545ba0ae53646c58a5a265aa5d535892a89b979fa4b1a49297a261a260555c5a.xhtml 3350] people cannot eat that contains wheat products and other foods with traces of gluten. Kumamolisin is an [http://partsregistry.org/wiki/index.php?title=Part:BBa_K590087 enzyme] that cleaves peptides and was produced by the iGEM-Team from the University of Washington] last year. The substrate includes a specific sequence of amino acids which causes celiac disease in sensitive people when they consume food containing gluten. Our beads could carry Kumamolisin and offer a protected passage through the stomach, so that the enzyme can work properly where it is needed in the intestines. The GerminationSTOP we put in place in our spores would ensure a correct dosage. This project is a pharmaceutical application and therefore would have to fulfill the laws for pharmaceuticals. This includes several verification steps of non-toxicity and efficacy.
CPX-Sporobeads: the relieve of the marine life
The excessive use of disposable plastic and the lack of universal recycling programs has led to the pollution of the world's oceans. In the ocean, large pieces of Polystyrene litter are ground by sea currents into very small pieces, so called plastik plankton, that are consumed by fish, filter feeders, and other organisms living in the oceans. Such plastic uptake can lead to poisoning, sterility and death. The [http://partsregistry.org/wiki/index.php/Part:BBa_I728500 CPX-peptide] generated by MIT (2007) can bind to Polystyrene. CPX-Sporobeads in huge filter boxes could be put into place to mechanically filter microscopic plastic particles, like polystyrene plankton, out of the water. To prevent the beads from being released into the sea and the plastic to be kept the Sporobeads could be attached to membranes in the boxes. The spores would then not only express CPX but also a membrane binding protein on their surface.
TALE-Sporobeads: easy and cheap detection of GMOs
Since 1990 green biotechnology releases many transgenic plants into the environment by selling genetically modified seeds. Thus organic farmers need to prove today, that their products meet the requirements for organic crops. Usually they pay laboratories to attest that this is the case. The new tools of molecular biology, TAL effectors, combined with our Sporobeads could be an easy and cheap solution for organic agriculture. Farmers could in future use our kit with TALE-lacZ-Sporobeads to detect genetically modified crops themselves. As spores are stable and safe vehicles, they could be send by mail without any considerations. The kit would be suitable for use outside of laboraty. The protocol for this could work as follows:
The DNA extracted from plants with solutions provided by the kit, is immobilized and fixed on a nitrocellulose membrane. This membrane is then washed, incubated with Sporobeads in solution and washed again. With addition of the substrate X-Gal, the lacZ of bound Sporobeads will catalyze the reaction so that a blue staining appears. If no such DNA is present, the spores will not bind and no blue color will appear.

| 
| 
| 
|
| Bacillus Intro | Bacillus BioBrickBox | Sporobeads | Germination STOP |
 "
"