Team:ZJU-China/project.htm
From 2012.igem.org
(Difference between revisions)
| Line 401: | Line 401: | ||
<img src="https://static.igem.org/mediawiki/igem.org/d/da/Zju_library_Fig1a.jpg" width="500px" /> | <img src="https://static.igem.org/mediawiki/igem.org/d/da/Zju_library_Fig1a.jpg" width="500px" /> | ||
<p>fig 1a. D0 is the original scaffold. D0 a-d were mutated to the scaffold with different aptamer arm length. </p> | <p>fig 1a. D0 is the original scaffold. D0 a-d were mutated to the scaffold with different aptamer arm length. </p> | ||
| - | <img src=" | + | <img src="hthttps://static.igem.org/mediawiki/igem.org/f/f7/Zju_library_fig1b.jpg" width="500px" /> |
<p>fig 1b. The result of arm length mutating. Both D0M4 and D0M5 scaffold half-on GEP.</p> | <p>fig 1b. The result of arm length mutating. Both D0M4 and D0M5 scaffold half-on GEP.</p> | ||
<p> </p> | <p> </p> | ||
| Line 462: | Line 462: | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<h2>1. Salicylate pathway</h2> | <h2>1. Salicylate pathway</h2> | ||
| - | < | + | <h2>(Group: iGEM2006_MIT)</h2> |
<img src="http://www.jiajunlu.com/igem/zju_iaa_1.jpg" width="600px" /> | <img src="http://www.jiajunlu.com/igem/zju_iaa_1.jpg" width="600px" /> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| Line 470: | Line 470: | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<h2>2. Pyocyanin pathway</h2> | <h2>2. Pyocyanin pathway</h2> | ||
| - | < | + | <h2>(Group: iGEM2007_Glasgow)</h2> |
<img src="http://www.jiajunlu.com/igem/zju_iaa_2.jpg" width="600px" /> | <img src="http://www.jiajunlu.com/igem/zju_iaa_2.jpg" width="600px" /> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| Line 478: | Line 478: | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<h2>3. Lycopene pathway</h2> | <h2>3. Lycopene pathway</h2> | ||
| - | < | + | <h2>(Group: iGEM2009_Cambridge) </h2> |
<img src="http://www.jiajunlu.com/igem/zju_iaa_3.jpg" width="600px" /> | <img src="http://www.jiajunlu.com/igem/zju_iaa_3.jpg" width="600px" /> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| Line 486: | Line 486: | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<h2>4. Holo-α-phycoerythrocyanin pathway</h2> | <h2>4. Holo-α-phycoerythrocyanin pathway</h2> | ||
| - | < | + | <h2>(Group: iGEM2004_UTAustin)</h2> |
<img src="http://www.jiajunlu.com/igem/zju_iaa_4.jpg" width="600px" /> | <img src="http://www.jiajunlu.com/igem/zju_iaa_4.jpg" width="600px" /> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| Line 494: | Line 494: | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<h2>5. BPA degradation pathway</h2> | <h2>5. BPA degradation pathway</h2> | ||
| - | < | + | <h2>(Group: iGEM2008_University_of_Alberta)</h2> |
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">Assessment: </p> | <p align="justify">Assessment: </p> | ||
| Line 501: | Line 501: | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<h2>6. IAM pathway</h2> | <h2>6. IAM pathway</h2> | ||
| - | < | + | <h2>(Group: iGEM2011_Imperial)</h2> |
<img src="http://www.jiajunlu.com/igem/zju_iaa_5.jpg" width="600px" /> | <img src="http://www.jiajunlu.com/igem/zju_iaa_5.jpg" width="600px" /> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
| Line 676: | Line 676: | ||
<!--Content Goes Here--> | <!--Content Goes Here--> | ||
| - | |||
<h2>1. Riboscaffold and targeted drug delivery therapy</h2> | <h2>1. Riboscaffold and targeted drug delivery therapy</h2> | ||
<p>This is an extension application of our designed clover series of riboscaffold. Some diseases, such as Cancer, will release some small molecular or change microenvironments beside it thus produce detectable signals. Different from using a biosensor to detect these signals, we utilize our scaffold’s aptamer, accompanying with the production of medicine target the disease. If we change Theophylline aptamer into nidus(disease) molecular aptamer, when riboscaffold bind nidus molecular and change conformation, MS2 aptamer & PP7 aptamer are going to set closer. Enzymes which combining MS2 aptamer & PP7 aptamer and producing drugs are ready to catalyze thus bring out targeting agents. It turns out to be a one-stop agency, once detect the focus of diease, will generate corresponding drug targeting the diease. </p> | <p>This is an extension application of our designed clover series of riboscaffold. Some diseases, such as Cancer, will release some small molecular or change microenvironments beside it thus produce detectable signals. Different from using a biosensor to detect these signals, we utilize our scaffold’s aptamer, accompanying with the production of medicine target the disease. If we change Theophylline aptamer into nidus(disease) molecular aptamer, when riboscaffold bind nidus molecular and change conformation, MS2 aptamer & PP7 aptamer are going to set closer. Enzymes which combining MS2 aptamer & PP7 aptamer and producing drugs are ready to catalyze thus bring out targeting agents. It turns out to be a one-stop agency, once detect the focus of diease, will generate corresponding drug targeting the diease. </p> | ||
| - | <img src= | + | <img src="https://static.igem.org/mediawiki/igem.org/d/da/ZJU_persp_1.png" width="600px" / > |
Figure1. Riboscaffold which can detect and treat diseases. </p> | Figure1. Riboscaffold which can detect and treat diseases. </p> | ||
| Line 686: | Line 685: | ||
<p>If we use these aptamer in replace of the theophylline aptamer on our riboscaffold, we can make the riboscaffold shining upon the binding of signal compounds mentioned above. This is a cool method to visualize the states, dynamics and localization of riboscaffold in the living cell. </p> | <p>If we use these aptamer in replace of the theophylline aptamer on our riboscaffold, we can make the riboscaffold shining upon the binding of signal compounds mentioned above. This is a cool method to visualize the states, dynamics and localization of riboscaffold in the living cell. </p> | ||
| - | <img src= | + | <img src="https://static.igem.org/mediawiki/igem.org/9/95/Zju_persp_2.png" width="600px" / > |
<p>Figure2. Aptamers that can shine upon binding. </p> | <p>Figure2. Aptamers that can shine upon binding. </p> | ||
| Line 693: | Line 692: | ||
<p>But how to obtain these LEGO riboscaffolds? Wachtveitlb[2] has reported a fantastic method to detect RNA-RNA interaction by introducing fluorophores like 1-ethynylpyrene into the 2-position of RNA adenosine. When two single-stranded RNAs with this fluorophore base pair with each other, the fluorescence spectrum changes and thus suggesting their interaction. So it is hopeful to find the desired riboscaffolds as LEGO bricks by selecting from the library! </p> | <p>But how to obtain these LEGO riboscaffolds? Wachtveitlb[2] has reported a fantastic method to detect RNA-RNA interaction by introducing fluorophores like 1-ethynylpyrene into the 2-position of RNA adenosine. When two single-stranded RNAs with this fluorophore base pair with each other, the fluorescence spectrum changes and thus suggesting their interaction. So it is hopeful to find the desired riboscaffolds as LEGO bricks by selecting from the library! </p> | ||
| - | <img src= | + | <img src="https://static.igem.org/mediawiki/igem.org/b/bc/Zju_persp_4.png" width="600px" / > |
<p> LEGO bricks.</p> | <p> LEGO bricks.</p> | ||
| - | <img src= | + | <img src="https://static.igem.org/mediawiki/igem.org/5/56/Zju_persp_5.png" width="600px" / > |
<p> Long scaffold that has multiple binding sites.</p> | <p> Long scaffold that has multiple binding sites.</p> | ||
| - | <img src= | + | <img src="https://static.igem.org/mediawiki/igem.org/e/ea/Zju_persp_6.png" width="600px" / > |
<p> A possible device built by LEGO riboscaffold.</p> | <p> A possible device built by LEGO riboscaffold.</p> | ||
| - | <img src= | + | <img src="https://static.igem.org/mediawiki/igem.org/8/81/Zju_presp_7.png" width="600px" / > |
<p> Sheets and tubes constructed by LEGO riboscaffolds in vivo.</p> | <p> Sheets and tubes constructed by LEGO riboscaffolds in vivo.</p> | ||
| Line 706: | Line 705: | ||
<p>Aptamers can be selected in vitro against nearly any target of choice. There are RNA aptamers that can specifically bind some transcriptional regulator. For example, Hunsicker [4] has selected one RNA aptamer that can bind TetR, which usually binds on operator sequence and repress gene expression. So once aptamers mentioned above are designed into a riboscaffold, it can initiate the expression of target genes with higher efficiency. </p> | <p>Aptamers can be selected in vitro against nearly any target of choice. There are RNA aptamers that can specifically bind some transcriptional regulator. For example, Hunsicker [4] has selected one RNA aptamer that can bind TetR, which usually binds on operator sequence and repress gene expression. So once aptamers mentioned above are designed into a riboscaffold, it can initiate the expression of target genes with higher efficiency. </p> | ||
| - | <img src= | + | <img src="https://static.igem.org/mediawiki/igem.org/1/15/Zju_persp_3.png" width="600px" / > |
<p>Figure3. Riboscaffold that can bring transcription factors to promoters. </p> | <p>Figure3. Riboscaffold that can bring transcription factors to promoters. </p> | ||
| Line 728: | Line 727: | ||
<p> [5] Kristina W. Thiel and Paloma H. Giangrande, Therapeutic Applications of DNA and RNA Aptamers. Oligonucleotides, 2009, Volume 19, Number 3, 209-222. </p> | <p> [5] Kristina W. Thiel and Paloma H. Giangrande, Therapeutic Applications of DNA and RNA Aptamers. Oligonucleotides, 2009, Volume 19, Number 3, 209-222. </p> | ||
<p> [6] Thodey, K. & Smolke, C.D. Bringing It Together with RNA. Science 333, 412-413 (2011). </p> | <p> [6] Thodey, K. & Smolke, C.D. Bringing It Together with RNA. Science 333, 412-413 (2011). </p> | ||
| + | |||
Revision as of 19:05, 26 September 2012

 "
"























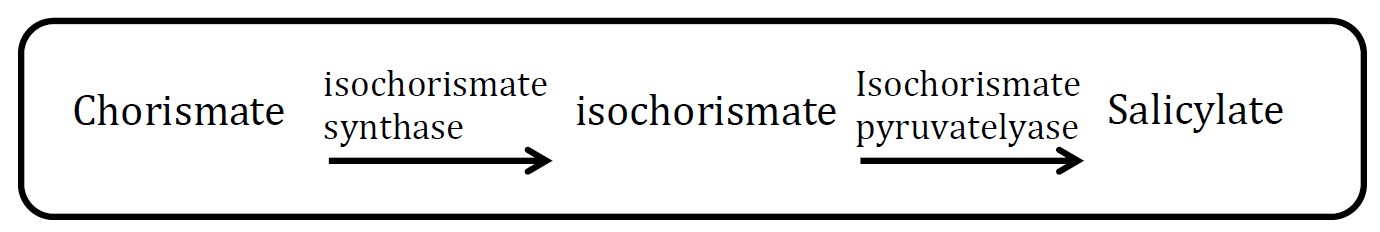
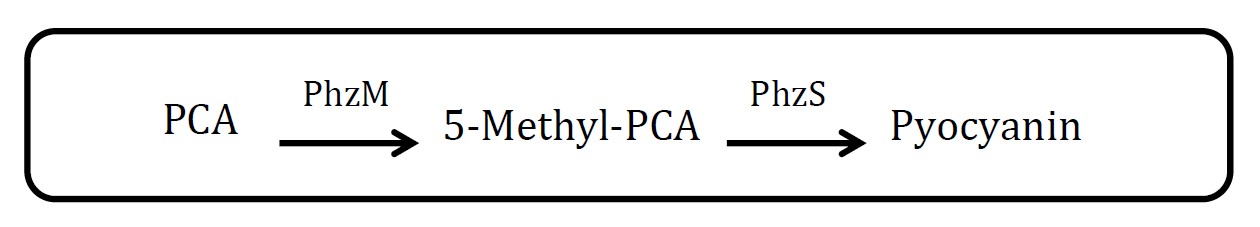
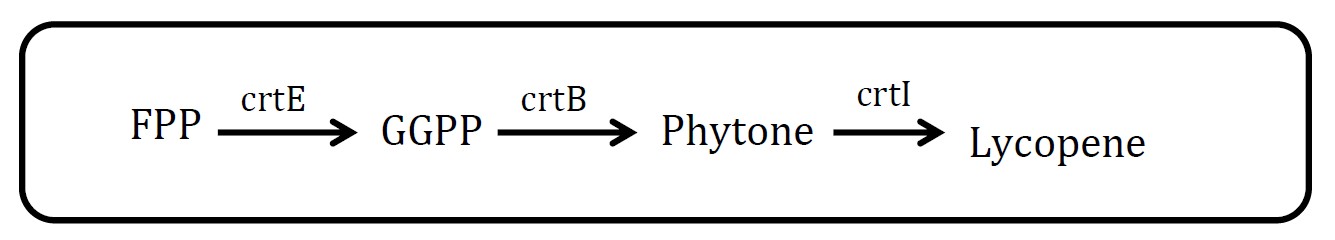
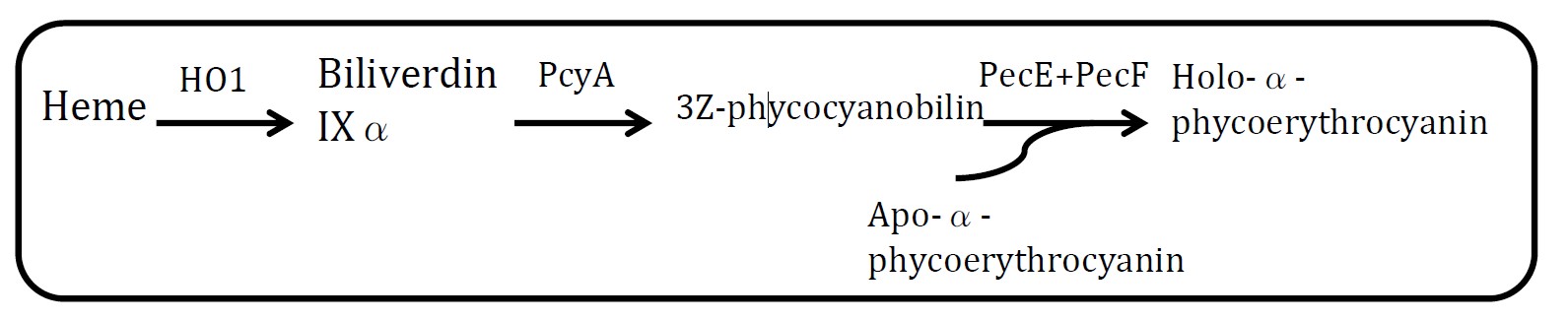
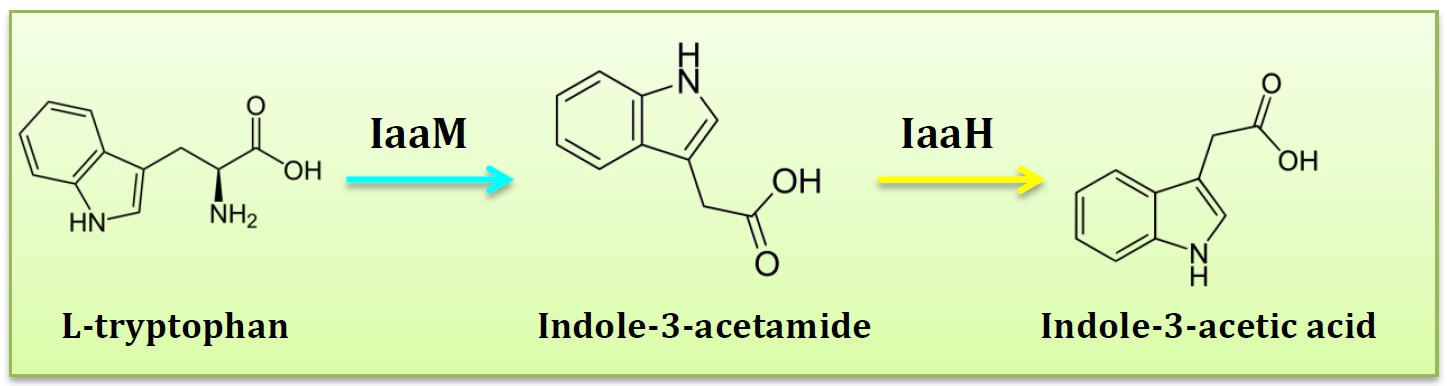









 Figure1. Riboscaffold which can detect and treat diseases.
Figure1. Riboscaffold which can detect and treat diseases.





