Team:SJTU-BioX-Shanghai/Project/project1.2
From 2012.igem.org
AleAlejandro (Talk | contribs) (→Fluorescence Complementation Assay) |
AleAlejandro (Talk | contribs) (→Fluorescence Complementation Assay) |
||
| Line 56: | Line 56: | ||
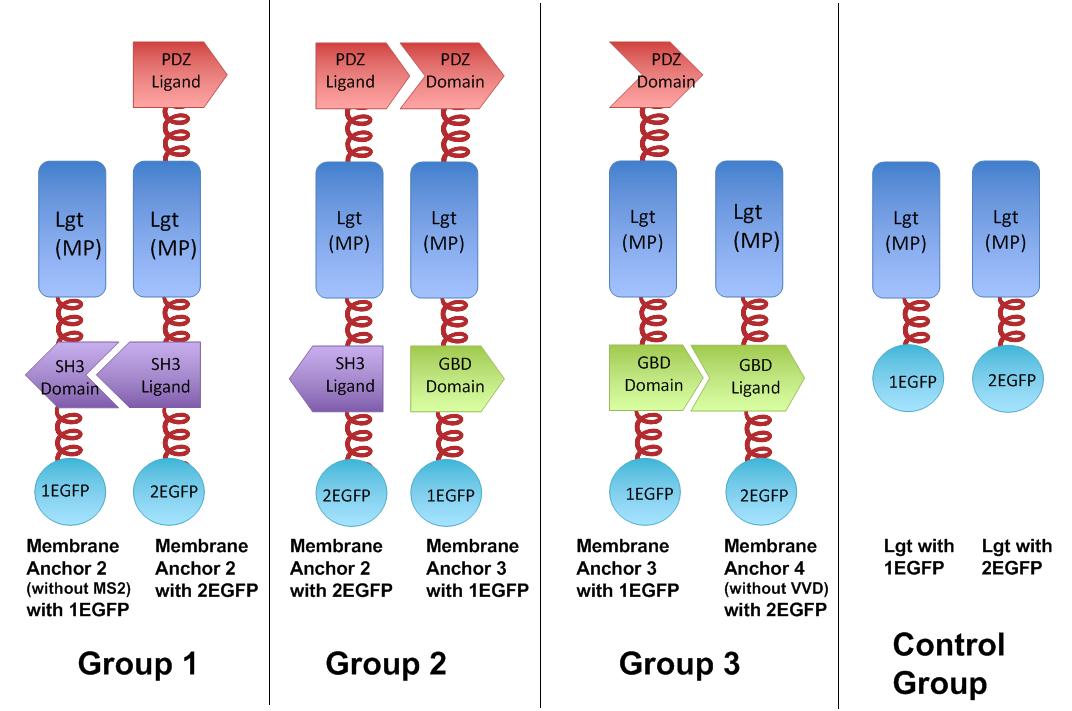
Proteins within each group are coexpressed in ''E.coli''. L-arabinose is not added into culture media until OD value of bacteria culture reaches 0.6. Bacteria are induced at L-Arabinose concentration of 0.2% for 6h. Bacteria samples are smeared onto glass slide and observed under laser confocal microscope. | Proteins within each group are coexpressed in ''E.coli''. L-arabinose is not added into culture media until OD value of bacteria culture reaches 0.6. Bacteria are induced at L-Arabinose concentration of 0.2% for 6h. Bacteria samples are smeared onto glass slide and observed under laser confocal microscope. | ||
| + | |||
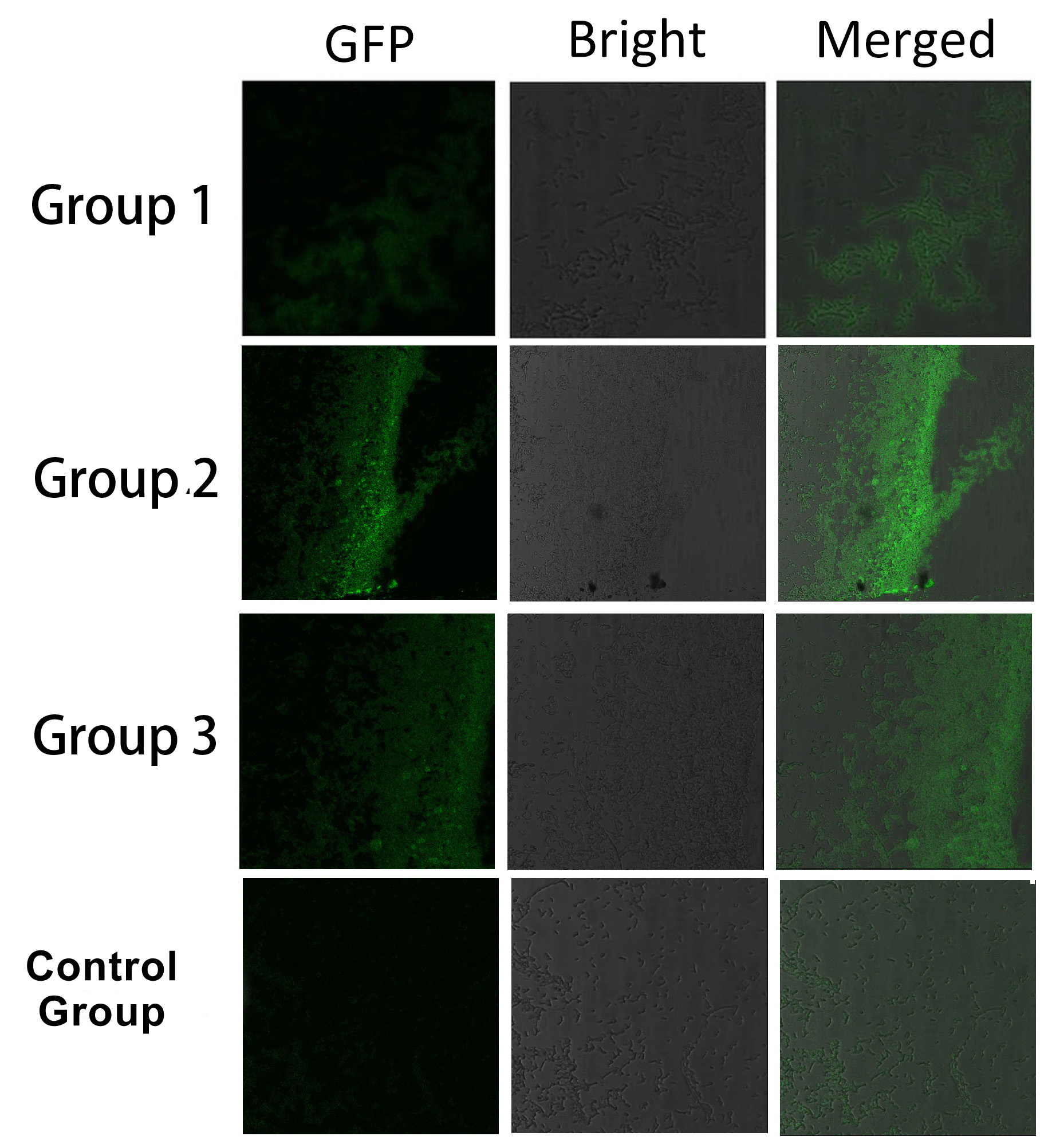
| + | [[Image:12SJTU_SplitEGFP.jpg|thumb|700px|center|''Fig.5'' :Bacteria in Group 1, 2 and 3 all show significantly stronger fluorescence intensity than Control Group. So we can conclude that through recruiting interacting domain and ligand, we can assemble proteins into a complex]] | ||
It is proved that membrane proteins with interacting proteins could interact and dimerize with each other. Thus, by recruiting ''E.coli'' native membrane protein Lgt, interacting proteins (SH3, PDZ, GBD) and downstream enzymes, we can easily build a ''Membrane Accelerator''. | It is proved that membrane proteins with interacting proteins could interact and dimerize with each other. Thus, by recruiting ''E.coli'' native membrane protein Lgt, interacting proteins (SH3, PDZ, GBD) and downstream enzymes, we can easily build a ''Membrane Accelerator''. | ||
Revision as of 19:20, 26 September 2012
| ||
|
 "
"