Team:Colombia/Project/Experiments/Aliivibrio and Streptomyces
From 2012.igem.org
(→Aim) |
|||
| Line 1: | Line 1: | ||
{{https://2012.igem.org/User:Tabima}} | {{https://2012.igem.org/User:Tabima}} | ||
| + | |||
| + | <div class="right_box"> | ||
==Aim== | ==Aim== | ||
| Line 130: | Line 132: | ||
<nowiki>[4]</nowiki> [http://mic.sgmjournals.org/content/154/2/373.full Colson, S., Wezel, G.P. van, Craig, M., Noens, E.E.E., Nothaft, H., Mommaas, A.M., Titgemeyer, F., Joris, B., and Rigali, S. (2008). The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in ''Streptomyces coelicolor''. '''Microbiology ''154,''''' 373–382.] | <nowiki>[4]</nowiki> [http://mic.sgmjournals.org/content/154/2/373.full Colson, S., Wezel, G.P. van, Craig, M., Noens, E.E.E., Nothaft, H., Mommaas, A.M., Titgemeyer, F., Joris, B., and Rigali, S. (2008). The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in ''Streptomyces coelicolor''. '''Microbiology ''154,''''' 373–382.] | ||
| + | |||
| + | </div> | ||
Revision as of 18:56, 26 September 2012
Template:Https://2012.igem.org/User:Tabima
Contents |
Aim
Our system heavily relies in the ability of our bacteria to sense environmental cues belonging to the pathogens we’re particularly concerned of.
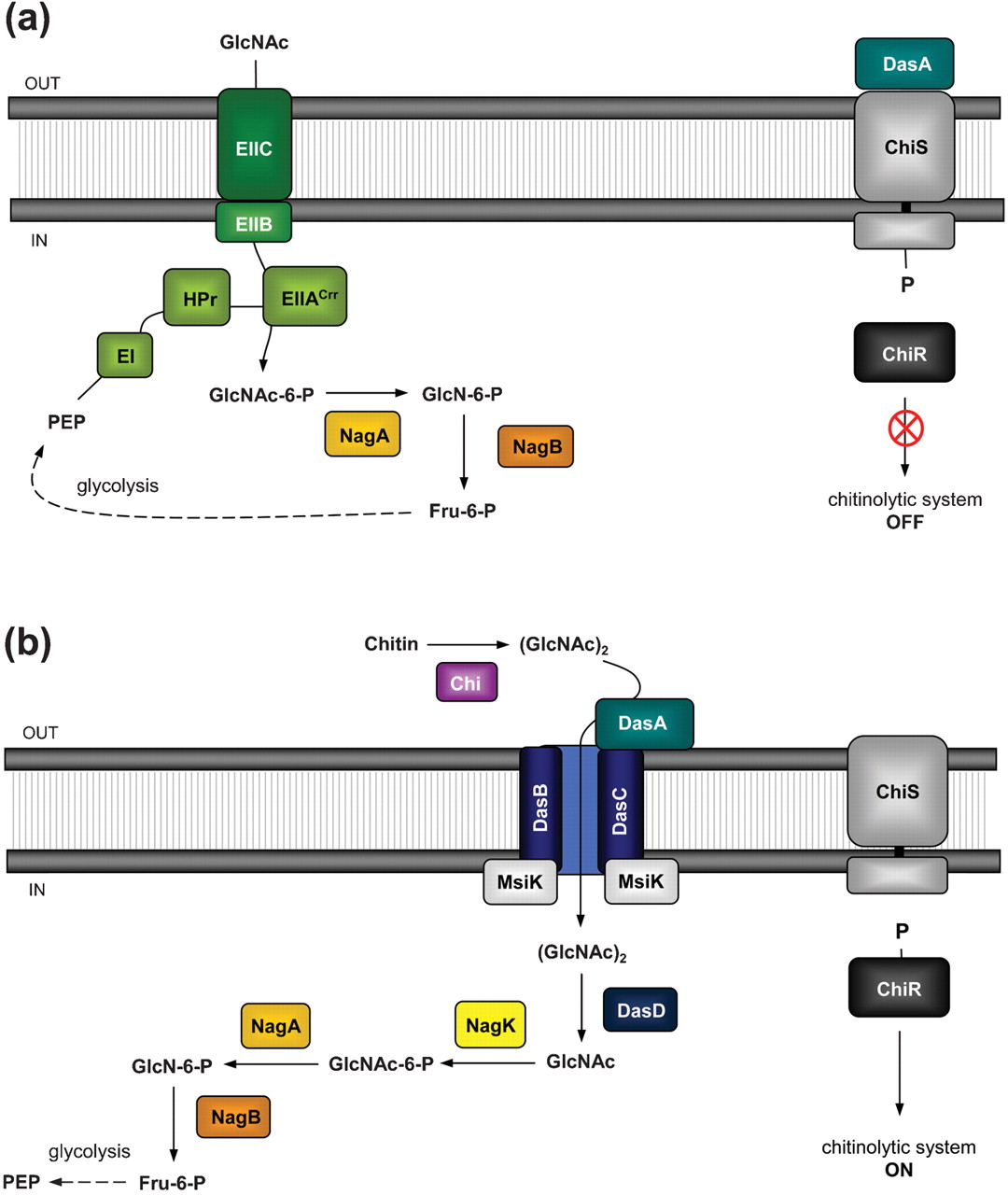
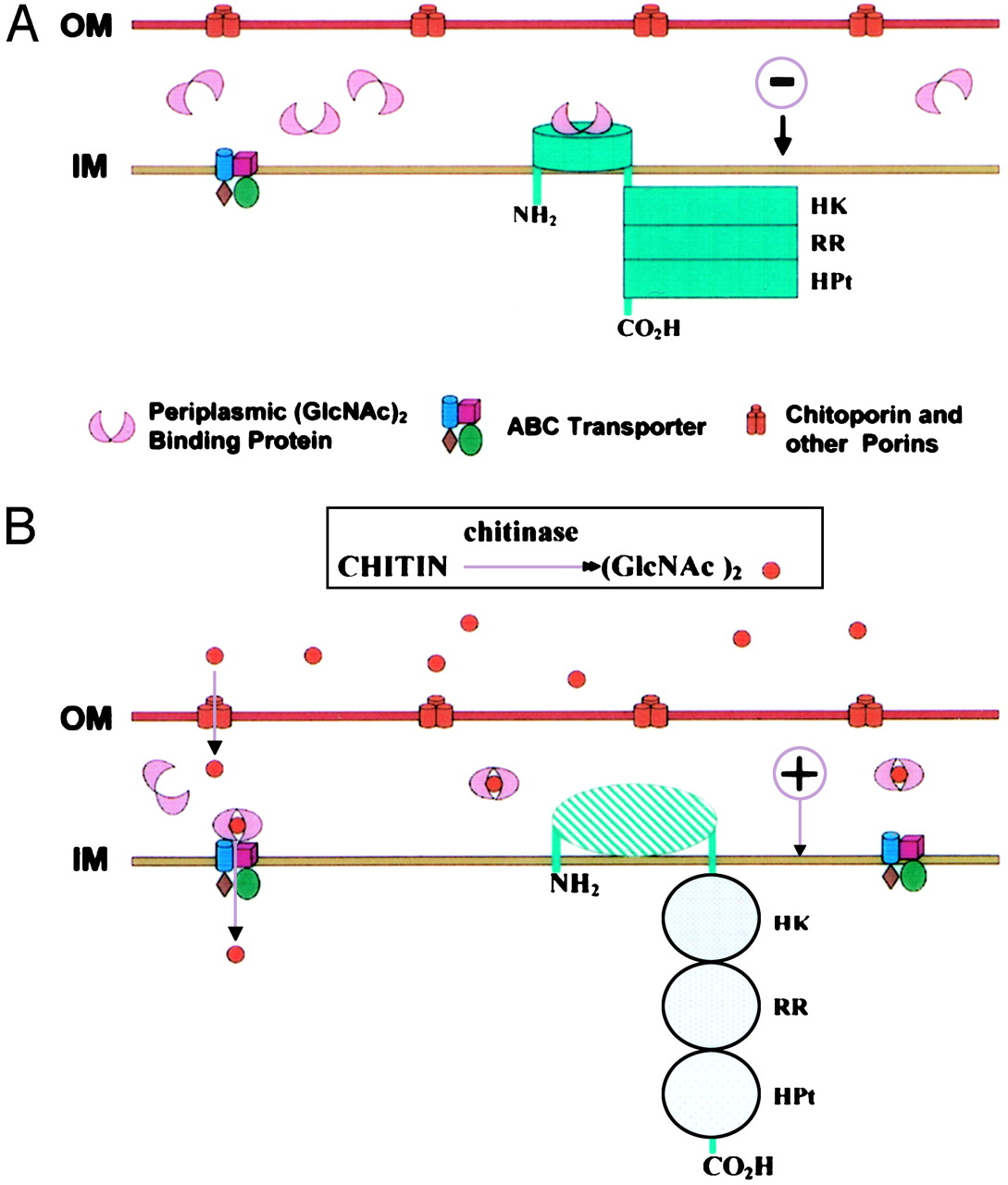
Chitin is one of the main components of fungal cell walls and, in consequence, we will use it as an indicator of fungal infection.
Vibrio fischeri ES114 ([http://ijs.sgmjournals.org/content/57/12/2823 now Aliivibrio fischeri ES114]) and Streptomyces coelicolor A3(2) are environmental bacteria with well-characterized detection and catabolic cascades for chitin use as a carbon source. The way by which each bacteria detect the chitin is through a two-component system. Figure 1 illustrates the proposed sensing model for the system in A. fischeri, whose activation depends on the interaction CBP + N-acetyl-glucosamine starting the signaling from ChiS histidine kinase to turn on the chitinolytic genes. Figure 2 depicts how S. coelicolor uses a pretty similar system to the previously described, to phosphorylate ChiR regulator and induce transcription of the catabolism genes.
In both cases, we expect to extract and put the genes into functional and independent biobricks available to further applications of chitin sensing and/or degradation.
Our bacteria
Aliivibrio fischeri ES114
| Aliivibrio fischeri ES114 | ||
|---|---|---|
| Taxonomy[1] | Superkingdom | Bacteria |
| Phylum | Proteobacteria | |
| Class | Gammaproteobacteria | |
| Order | Vibrionales | |
| Family | Vibrionaceae | |
| Genus | Aliivibrio | |
| Species | A. fischeri | |
| Strain | ES114 | |
Streptomyces coelicolor A3(2)
| Streptomyces coelicolor A3(2) | ||
|---|---|---|
| Taxonomy[2] | Superkingdom | Bacteria |
| Phylum | Actinobacteria | |
| Class | Actinobacteria | |
| Order | Actinomycetales | |
| Family | Streptomycetaceae | |
| Genus | Streptomyces | |
| Species | Streptomyces coelicolor | |
| Strain | A3(2) | |
References
[1] [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=312309&lvl=3&lin=f&keep=1&srchmode=1&unlock NCBI Taxonomy Browser: Aliivibrio fischeri ES114]
[2] [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=312309&lvl=3&lin=f&keep=1&srchmode=1&unlock NCBI Taxonomy Browser: Streptomyces coelicolor A3(2)]
[3] [http://www.pnas.org/content/101/2/627 Li, X., and Roseman, S. (2004). The chitinolytic cascade in Vibrios is regulated by chitin oligosaccharides and a two-component chitin catabolic sensor/kinase. PNAS 101, 627–631.]
[4] [http://mic.sgmjournals.org/content/154/2/373.full Colson, S., Wezel, G.P. van, Craig, M., Noens, E.E.E., Nothaft, H., Mommaas, A.M., Titgemeyer, F., Joris, B., and Rigali, S. (2008). The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154, 373–382.]
 "
"