Team:Marburg SYNMIKRO/Notebook
From 2012.igem.org
(Difference between revisions)
Enantiomere (Talk | contribs) |
Enantiomere (Talk | contribs) |
||
| Line 163: | Line 163: | ||
==18.07.2012== | ==18.07.2012== | ||
; Agarose gel | ; Agarose gel | ||
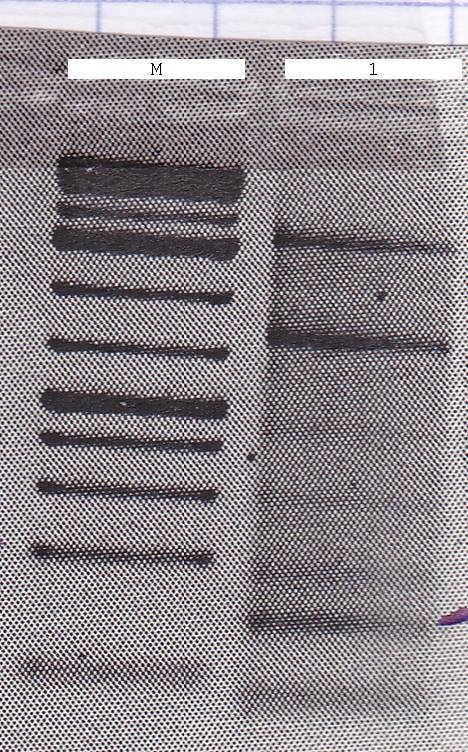
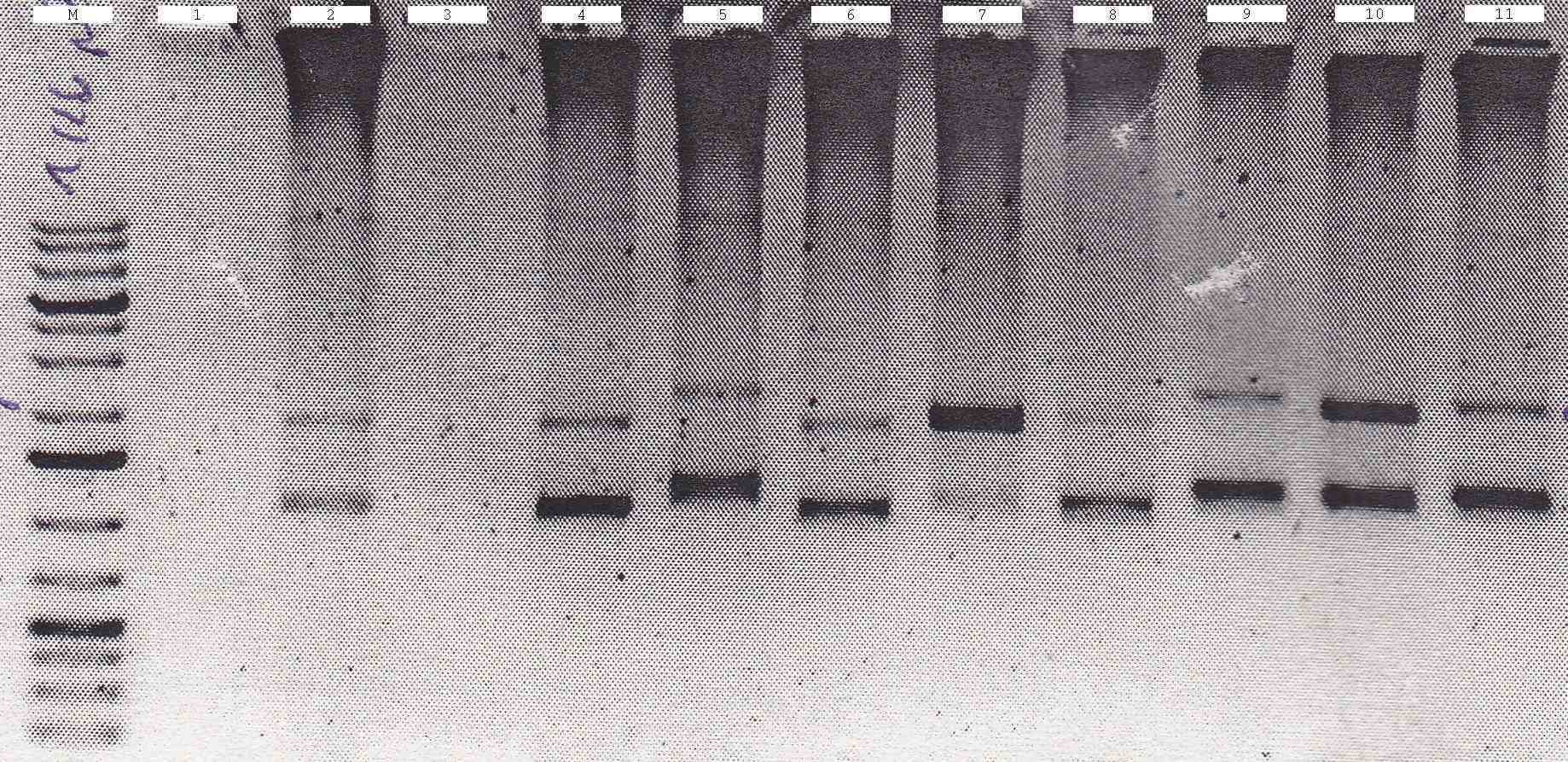
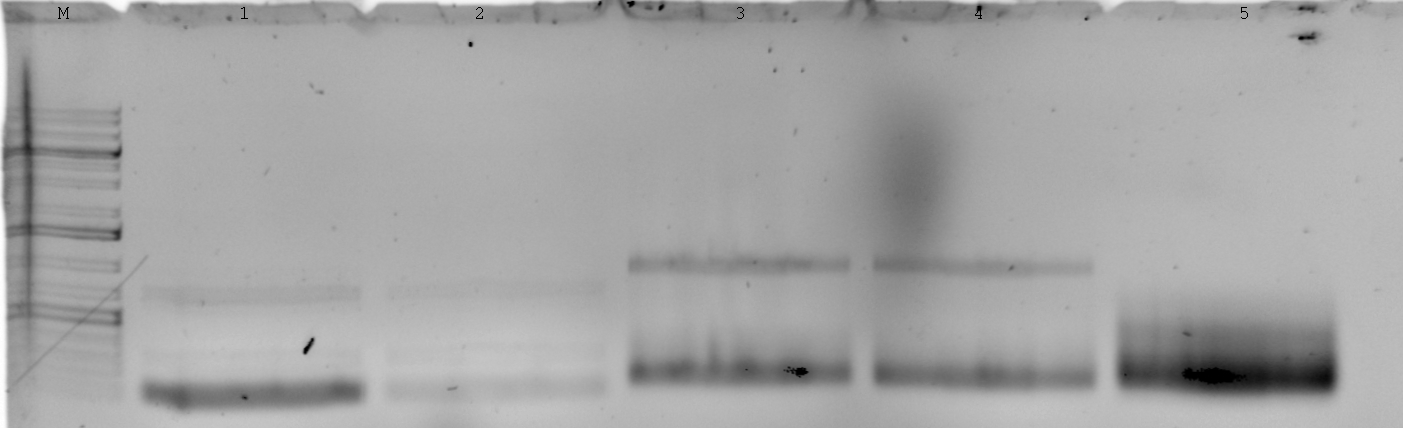
| - | [[File:Syn_MR_Gel1.jpg|thumb|800px|center|M:1kb gene ruler plus, 1:enhancer, 2:GroES, 3:AmiC, 4:HU, 5:GixR, 6:GFP, 7:GFP-Fusion, 8:CFP, 9:mRFP, 10:RBS, 11:LacIQ, 12:P108, 13:P100, 14: | + | [[File:Syn_MR_Gel1.jpg|thumb|800px|center|M:1kb gene ruler plus, 1:enhancer, 2:GroES, 3:AmiC, 4:HU, 5:GixR, 6:GFP, 7:GFP-Fusion, 8:CFP, 9:mRFP, 10:RBS, 11:LacIQ, 12:P108, 13:P100, 14:?-DNA]] |
; DNA Gel extraction | ; DNA Gel extraction | ||
| Line 184: | Line 184: | ||
; PCR | ; PCR | ||
| - | : AmiC (primer: MS1 + MS2) | + | : AmiC (primer: [[Team:Marburg_SYNMIKRO/Primer#MS1|MS1]] + [[Team:Marburg_SYNMIKRO/Primer#MS2|MS2]]) |
| Line 252: | Line 252: | ||
; PCR | ; PCR | ||
| - | : CFP (primer: MS16 + MS17) ⇒ template-DNA: BBa_E0010 | + | : CFP (primer: [[Team:Marburg_SYNMIKRO/Primer#MS16|MS16]] + [[Team:Marburg_SYNMIKRO/Primer#MS17|MS17]]) ⇒ template-DNA: BBa_E0010 |
: ⇒ positive | : ⇒ positive | ||
| - | : mRFP (primer: MS14 + MS15) ⇒ template-DNA: BBa_E1010 | + | : mRFP (primer: [[Team:Marburg_SYNMIKRO/Primer#MS14|MS14]] + [[Team:Marburg_SYNMIKRO/Primer#MS15|MS15]]) ⇒ template-DNA: BBa_E1010 |
: ⇒ positive | : ⇒ positive | ||
| - | : Gin (primer: MS11 + MS13) ⇒ template-DNA: chromosomal DNA (that was wrong) | + | : Gin (primer: [[Team:Marburg_SYNMIKRO/Primer#MS11|MS11]] + [[Team:Marburg_SYNMIKRO/Primer#MS13|MS13]]) ⇒ template-DNA: chromosomal DNA (that was wrong) |
| - | : RBS-Gin (primer: MS13 + MS12) ⇒ template-DNA: chromosomal DNA (that was wrong) | + | : RBS-Gin (primer: [[Team:Marburg_SYNMIKRO/Primer#MS13|MS13]] + [[Team:Marburg_SYNMIKRO/Primer#MS12|MS12]]) ⇒ template-DNA: chromosomal DNA (that was wrong) |
: ⇒ annealing: 30 sec; 60 °C, elongation: 40 sec; 72 °C, 35 cycles | : ⇒ annealing: 30 sec; 60 °C, elongation: 40 sec; 72 °C, 35 cycles | ||
| Line 295: | Line 295: | ||
; PCR | ; PCR | ||
| - | : AmiC (primer: MS1 + MS2) | + | : AmiC (primer: [[Team:Marburg_SYNMIKRO/Primer#MS1|MS1]] + [[Team:Marburg_SYNMIKRO/Primer#MS2|MS2]]) |
| - | : RBS-Gin (primer: MS12 + MS13) | + | : RBS-Gin (primer: [[Team:Marburg_SYNMIKRO/Primer#MS12|MS12]] + [[Team:Marburg_SYNMIKRO/Primer#MS13|MS13]]) |
| - | : Gin (primer: MS11 + MS13) | + | : Gin (primer: [[Team:Marburg_SYNMIKRO/Primer#MS11|MS11]] + [[Team:Marburg_SYNMIKRO/Primer#MS13|MS13]]) |
: ⇒ template-DNA: chromosomal DNA, annealing: 30 sec; 55 °C, elongation: 40 sec; 72 °C, 35 cycles | : ⇒ template-DNA: chromosomal DNA, annealing: 30 sec; 55 °C, elongation: 40 sec; 72 °C, 35 cycles | ||
| Line 615: | Line 615: | ||
; PCR | ; PCR | ||
| - | : GroES (primer: MS5 + | + | : GroES (primer: [[Team:Marburg_SYNMIKRO/Primer#MS5|MS5]] + [[Team:Marburg_SYNMIKRO/Primer#MS6b|MS6b]]) |
| - | : HU (primer: MS3 + | + | : HU (primer: [[Team:Marburg_SYNMIKRO/Primer#MS3|MS3]] + [[Team:Marburg_SYNMIKRO/Primer#MS4b|MS4b]]) |
: ⇒ with and without DMSO, template-DNA: chromosomal DNA, annealing: 30 sec; 60 °C, elongation: 40 sec; 72 °C, 35 cycles | : ⇒ with and without DMSO, template-DNA: chromosomal DNA, annealing: 30 sec; 60 °C, elongation: 40 sec; 72 °C, 35 cycles | ||
| Line 654: | Line 654: | ||
; PCR | ; PCR | ||
| - | : GroES (primer: MS5 + | + | : GroES (primer: [[Team:Marburg_SYNMIKRO/Primer#MS5|MS5]] + [[Team:Marburg_SYNMIKRO/Primer#MS6b|MS6b]]) |
| - | : HU (primer: MS3 + | + | : HU (primer: [[Team:Marburg_SYNMIKRO/Primer#MS3|MS3]] + [[Team:Marburg_SYNMIKRO/Primer#MS4b|MS4b]]) |
: ⇒ with and without DMSO, template-DNA: chromosomal DNA, annealing: 30 sec; 55 °C, elongation: 30 sec; 72 °C, 35 cycles | : ⇒ with and without DMSO, template-DNA: chromosomal DNA, annealing: 30 sec; 55 °C, elongation: 30 sec; 72 °C, 35 cycles | ||
| Line 688: | Line 688: | ||
; PCR | ; PCR | ||
| - | : GroES (primer: MS5 + | + | : GroES (primer: [[Team:Marburg_SYNMIKRO/Primer#MS5|MS5]] + [[Team:Marburg_SYNMIKRO/Primer#MS6b|MS6b]]) |
: ⇒ with and without DMSO, template-DNA: chromosomal DNA, annealing: 30 sec; 55 °C, elongation: 30 sec; 72 °C , 35 cycles | : ⇒ with and without DMSO, template-DNA: chromosomal DNA, annealing: 30 sec; 55 °C, elongation: 30 sec; 72 °C , 35 cycles | ||
| Line 781: | Line 781: | ||
; PCR | ; PCR | ||
| - | : Backbone (primer MS24 + MS25) | + | : Backbone (primer [[Team:Marburg_SYNMIKRO/Primer#MS24|MS24]] + [[Team:Marburg_SYNMIKRO/Primer#MS25|MS25]]) |
: ⇒ template-DNA: pSB1A3, pSB1T3, pSB1C3 and pSB1K3, annealing: 30 sec; 55 °C, elongation: 30 sec; 72 °C , 35 cycles | : ⇒ template-DNA: pSB1A3, pSB1T3, pSB1C3 and pSB1K3, annealing: 30 sec; 55 °C, elongation: 30 sec; 72 °C , 35 cycles | ||
| Line 810: | Line 810: | ||
: pSB1C3-BBa_J04450 (template DNA) | : pSB1C3-BBa_J04450 (template DNA) | ||
: pSB1K3-BBa_J04450 (template DNA) | : pSB1K3-BBa_J04450 (template DNA) | ||
| - | : used primer --> | + | : used primer --> [[Team:Marburg_SYNMIKRO/Primer#MS24|MS24]] + [[Team:Marburg_SYNMIKRO/Primer#MS25|MS25]] |
: initial denaturation 98°C - 5min | : initial denaturation 98°C - 5min | ||
: denaturation: 98°C - 1min; annealing: 55°C - 30s; elongation: 72°C - 2.5min -->repeat 35 times | : denaturation: 98°C - 1min; annealing: 55°C - 30s; elongation: 72°C - 2.5min -->repeat 35 times | ||
| - | : 72°C - 5min --> 4°C - | + | : 72°C - 5min --> 4°C - 8 |
; miniprep | ; miniprep | ||
| Line 833: | Line 833: | ||
; PCR | ; PCR | ||
| - | : CFP | + | : CFP ([[Team:Marburg_SYNMIKRO/Primer#MS16|MS16]] + [[Team:Marburg_SYNMIKRO/Primer#MS17|MS17]]) |
| - | : mRFP | + | : mRFP ([[Team:Marburg_SYNMIKRO/Primer#MS14|MS14]] + [[Team:Marburg_SYNMIKRO/Primer#MS15|MS15]]) |
: initial denaturation 98°C - 5min | : initial denaturation 98°C - 5min | ||
: denaturation: 98°C - 10s; annealing: 60°C - 30s; elongation: 72°C - 40s -->repeat 35 times | : denaturation: 98°C - 10s; annealing: 60°C - 30s; elongation: 72°C - 40s -->repeat 35 times | ||
| - | : 72°C - 5min --> 4°C - | + | : 72°C - 5min --> 4°C - 8 |
; 1% Agarose gel | ; 1% Agarose gel | ||
| Line 871: | Line 871: | ||
: pSB1K3-BBa_J04450 (template DNA) | : pSB1K3-BBa_J04450 (template DNA) | ||
: pSB1T3-BBa_J04450 (template DNA) | : pSB1T3-BBa_J04450 (template DNA) | ||
| - | : used primer --> | + | : used primer --> [[Team:Marburg_SYNMIKRO/Primer#MS24|MS24]] + [[Team:Marburg_SYNMIKRO/Primer#MS25|MS25]] |
: initial denaturation 98°C - 5min | : initial denaturation 98°C - 5min | ||
: denaturation: 98°C - 1min; annealing: 55°C - 30s; elongation: 72°C - 2.5min -->repeat 35 times | : denaturation: 98°C - 1min; annealing: 55°C - 30s; elongation: 72°C - 2.5min -->repeat 35 times | ||
| - | : 72°C - 5min --> 4°C - | + | : 72°C - 5min --> 4°C - 8 |
; 1% Agarose gel | ; 1% Agarose gel | ||
| Line 898: | Line 898: | ||
: initial denaturation 98°C - 5min | : initial denaturation 98°C - 5min | ||
: denaturation: 98°C - 10s; annealing: 60°C - 30s; elongation: 72°C - 40s -->repeat 30 times | : denaturation: 98°C - 10s; annealing: 60°C - 30s; elongation: 72°C - 40s -->repeat 30 times | ||
| - | : 72°C - 10min --> 4°C - | + | : 72°C - 10min --> 4°C - 8 |
| - | : primer: MS22 | + | : primer: [[Team:Marburg_SYNMIKRO/Primer#MS22|MS22]] + [[Team:Marburg_SYNMIKRO/Primer#MS23|MS23]] |
; 1% Agarose gel | ; 1% Agarose gel | ||
| Line 937: | Line 937: | ||
; PCR | ; PCR | ||
| - | : template: backbones (pSB1K3/T3/C3/A3 | + | : template: backbones (pSB1K3/T3/C3/A3) |
| - | : parameters: 98°C 5min, >> 98°C 1min, 70°C 30s, 72°C 2.5min << x30, 72°C 5min, 4°C | + | : parameters: 98°C 5min, >> 98°C 1min, 70°C 30s, 72°C 2.5min << x30, 72°C 5min, 4°C 8 |
| + | : primer: [[Team:Marburg_SYNMIKRO/Primer#MS22|MS22]] + [[Team:Marburg_SYNMIKRO/Primer#MS23|MS23]] | ||
; colony-PCR | ; colony-PCR | ||
| - | : parameters: 98°C 5min, >> 98°C 1min, 72°C 1min << x30, 72°C 5min, 4°C | + | : parameters: 98°C 5min, >> 98°C 1min, 72°C 1min << x30, 72°C 5min, 4°C 8 |
| + | : primer: [[Team:Marburg_SYNMIKRO/Primer#MS18|MS18]] + [[Team:Marburg_SYNMIKRO/Primer#MS19|MS19]] | ||
; Agarose gel of backbone PCR | ; Agarose gel of backbone PCR | ||
| Line 970: | Line 972: | ||
: Enhancer, Terminator, Gix, P-RBS | : Enhancer, Terminator, Gix, P-RBS | ||
: parameters: 98°C 5', 98°C 1', 66°C 30", 72°C 30", 72°C 5', 4°C ~ | : parameters: 98°C 5', 98°C 1', 66°C 30", 72°C 30", 72°C 5', 4°C ~ | ||
| + | : primer: [[Team:Marburg_SYNMIKRO/Primer#MS20|MS20]] + [[Team:Marburg_SYNMIKRO/Primer#MS21|MS21]] | ||
; 1% Agarose gele | ; 1% Agarose gele | ||
| Line 993: | Line 996: | ||
: diluted templates: Terminator, P-RBS, Gix 1 --> 1:10 | : diluted templates: Terminator, P-RBS, Gix 1 --> 1:10 | ||
: Enhancer, Gix 2 --> 1:20 | : Enhancer, Gix 2 --> 1:20 | ||
| + | : parameters: 98°C 5', 98°C 1', 66°C 30", 72°C 30", 72°C 5', 4°C ~ | ||
| + | : primer: [[Team:Marburg_SYNMIKRO/Primer#MS20|MS20]] + [[Team:Marburg_SYNMIKRO/Primer#MS21|MS21]] | ||
| + | |||
; 1% Agarose gele | ; 1% Agarose gele | ||
| Line 1,049: | Line 1,055: | ||
: Parameters No.1: 95°C 5', >>95°C 30", 60°C 30", 72°C 1'<< x30, 72°C 5', 4°C | : Parameters No.1: 95°C 5', >>95°C 30", 60°C 30", 72°C 1'<< x30, 72°C 5', 4°C | ||
: Parameters No.2: 95°C 5', >>95°C 30", 72°C 1.5'<< x30, 72°C 5', 4°C | : Parameters No.2: 95°C 5', >>95°C 30", 72°C 1.5'<< x30, 72°C 5', 4°C | ||
| + | : Primer [[Team:Marburg_SYNMIKRO/Primer#MS26|MS26]] + [[Team:Marburg_SYNMIKRO/Primer#MS27|MS27]] | ||
| + | |||
; 1% Agarose gele | ; 1% Agarose gele | ||
| Line 1,110: | Line 1,118: | ||
: of colonies (CFP-T, mRFP-T) | : of colonies (CFP-T, mRFP-T) | ||
: parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C | : parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C | ||
| + | : primer [[Team:Marburg_SYNMIKRO/Primer#MS18|MS18]] + [[Team:Marburg_SYNMIKRO/Primer#MS19|MS19]] | ||
; Agarose gel | ; Agarose gel | ||
| Line 1,145: | Line 1,154: | ||
:AmiC-GFP ⇒; no fluorescence | :AmiC-GFP ⇒; no fluorescence | ||
| - | ;colony-PCR | + | ; colony-PCR |
| - | :GixR | + | : GixR |
| - | :RBS-Gin | + | : RBS-Gin |
| - | :Terminator | + | : Terminator |
| + | : parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C | ||
| + | : primer [[Team:Marburg_SYNMIKRO/Primer#MS20|MS20]] + [[Team:Marburg_SYNMIKRO/Primer#MS21|MS21]] | ||
;3A-Assembly (digestion, ligation, transformation) | ;3A-Assembly (digestion, ligation, transformation) | ||
| Line 1,177: | Line 1,188: | ||
: RBS (B0030) | : RBS (B0030) | ||
: P-RBS (108) | : P-RBS (108) | ||
| + | : parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C | ||
| + | : primer [[Team:Marburg_SYNMIKRO/Primer#MS20|MS20]] + [[Team:Marburg_SYNMIKRO/Primer#MS21|MS21]] | ||
; Agarose gel | ; Agarose gel | ||
| Line 1,225: | Line 1,238: | ||
::: -one using GC-Buffer | ::: -one using GC-Buffer | ||
::: -and another one using self-made reaction buffer | ::: -and another one using self-made reaction buffer | ||
| - | : Primer used : MS12 | + | : Primer used : [[Team:Marburg_SYNMIKRO/Primer#MS12|MS12]] + [[Team:Marburg_SYNMIKRO/Primer#MS13|MS13]] |
: Template DNA : ''E. coli'' C600 chromosomal DNA | : Template DNA : ''E. coli'' C600 chromosomal DNA | ||
| Line 1,237: | Line 1,250: | ||
; colony-PCR | ; colony-PCR | ||
: AmiC-T & GroES-T (transformation 11.09.12) | : AmiC-T & GroES-T (transformation 11.09.12) | ||
| - | + | : parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C | |
| + | : primer [[Team:Marburg_SYNMIKRO/Primer#MS20|MS20]] + [[Team:Marburg_SYNMIKRO/Primer#MS21|MS21]] | ||
; 1% Agarose gele | ; 1% Agarose gele | ||
| Line 1,374: | Line 1,388: | ||
: RBS-Gin | : RBS-Gin | ||
: 2x Terminator | : 2x Terminator | ||
| + | : parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C | ||
| + | : primer [[Team:Marburg_SYNMIKRO/Primer#MS20|MS20]] + [[Team:Marburg_SYNMIKRO/Primer#MS21|MS21]] | ||
; 1% Agarose gele | ; 1% Agarose gele | ||
| Line 1,417: | Line 1,433: | ||
: RBS-Gin (from 17.09.12) | : RBS-Gin (from 17.09.12) | ||
: 2x Terminator B0017 (from 17.09.12) | : 2x Terminator B0017 (from 17.09.12) | ||
| + | : parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C | ||
| + | : primer [[Team:Marburg_SYNMIKRO/Primer#MS20|MS20]] + [[Team:Marburg_SYNMIKRO/Primer#MS21|MS21]] | ||
: --> Amplification failed | : --> Amplification failed | ||
Revision as of 17:30, 26 September 2012

Contents |
Week 1
[all]
09.07. - 15.07.2012
09.07.2012
- Inoculation
- E. coli strain C600 Mucts62 in dYT-medium
10.07.2012
- Isolation of chromosomal DNA
- E. coli strain C600 Mucts62 (lysogenic for temperature sensitive phage Mu cts62)
- Inoculation
- E. coli strain DH5α and TOP10 in dYT-medium
11.07.2012
- Preparation of chemically competent E. coli cells
- TOP10
12.07.2012
- Transformation
- GFP, GFP_fusion, CFP, mRFP, RBS, P108, P100, Plac with E.coli TOP10
| Name | Biobrick | Plate | Well | Backbone | Resistance |
|---|---|---|---|---|---|
| GFP | [http://partsregistry.org/Part:BBa_E0040 BBa_E0040] | 1 | 14K | pSB1A2 | Ampicillin |
| GFP_fusion | [http://partsregistry.org/Part:BBa_K125500 BBa_K125500] | 3 | 2P | pSB1A2 | Ampicillin |
| CFP | [http://partsregistry.org/Part:BBa_E0020 BBa_E0020] | 1 | 6A | pSB1A2 | Ampicillin |
| mRFP | [http://partsregistry.org/Part:BBa_E1010 BBa_E1010] | 1 | 18F | pSB2K3 | Kanamycin |
| RBS | [http://partsregistry.org/Part:BBa_J61100 BBa_J61100] | 1 | 5J | pSB1A2 | Ampicillin |
| P108 | [http://partsregistry.org/Part:BBa_J23108 BBa_J23108] | 2 | 2E | BBa_J61002 | Ampicillin |
| P100 | [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] | 1 | 18C | BBa_J61002 | Ampicillin |
| PLac | [http://partsregistry.org/Part:BBa_I14032 BBa_I14032] | 2 | 11P | pSB2K3 | Kanamycin |
Week 2
16.07. - 22.07.2012
16.07.2012
- PCR
- AmiC (primer: MS1 + MS2)
- HU (primer: MS3 + MS4)
- GroES (primer: MS5 + MS6)
- GixR (primer: MS7 + MS8)
- Enhancer (primer: MS9 + MS10)
- ⇒ template-DNA: chromosomal DNA, denaturation: 30 sec at 95°C; annealing: 30 sec at 60°C; elongation: 30 sec at 72°C; 35 cycles
- Inoculation
- GFP
- GFP_fusion
- CFP
- mRFP
- RBS
- P108
- LacIQ
- P100
17.07.2012
- Plasmid preparation
- GFP
- GFP_fusion
- CFP
- mRFP
- RBS
- P108
LacIq- P100
18.07.2012
- Agarose gel
- DNA Gel extraction
- Enhancer
- GroES
- HU
- GixR Hybridization
- 5 µl primer MS7 (100 µM) + 5 µl primer MS8 (100 µM)
- 5 min 99°C
- over night, room temperature
- -80°C
19.07.2012
- Fill-in reaction (primer extension)
- Enhancer
- Restriction
- Enhancer (EcoRI/PstI)
- ⇒ template-DNA: chromosomal DNA, annealing: 30 sec; 85 °C, elongation: 36 sec; 72 °C, 35 cycles, 1) 2 µl DMSO; 1 µl chrom. DNA | 2) 2 µl DMSO; 5 µl chrom. DNA | 3) without DMSO; 1 µl chrom. DNA | 4) without DMSO; 5 µl chrom. DNA | 5) 2 µl DMSO; without chrom. DNA
- Test restriction digest
- 7) CFP (EcoRI/PstI)
- 8) GFP (EcoRI/PstI)
- 9) GFP-fusion (EcoRI/PstI)
- 10) mRFP (EcoRI/PstI)
- ⇒ all negative
- Agarose gel
- Control digest
- CFP (EcoRI/PstI)
- GFP (EcoRI/PstI)
- GFP-fusion (EcoRI/PstI)
- mRFP (EcoRI/PstI)
- ⇒ restriction over night
20.07.2012
- Agarose gel from the control digest
Week 3
23.07. - 29.07.2012
26.07.2012
- Restriction
- pSB1T3 (XbaI/SpeI and DpnI)
- pSB1A3 (XbaI/SpeI and DpnI)
- pSB1K3 (XbaI/SpeI and DpnI)
- pSB1C3 (XbaI/SpeI and DpnI)
- Ligation
- GixR + pSB1A3
- Enhancer + pSB1K3
- cyclisation of pSB1C3, pSB1K3, pSB1A3, pSB1T3 (couldn’t work)
Week 4
30.07. - 05.08.2012
30.07.2012
- PCR
- CFP (primer: MS16 + MS17) ⇒ template-DNA: BBa_E0010
- ⇒ positive
- mRFP (primer: MS14 + MS15) ⇒ template-DNA: BBa_E1010
- ⇒ positive
- Gin (primer: MS11 + MS13) ⇒ template-DNA: chromosomal DNA (that was wrong)
- RBS-Gin (primer: MS13 + MS12) ⇒ template-DNA: chromosomal DNA (that was wrong)
- ⇒ annealing: 30 sec; 60 °C, elongation: 40 sec; 72 °C, 35 cycles
- Transformation
- all vector-backbone-ligations
- Enhancer (K3)
- GixR (A3)
31.07.2012
- Result
- transformation
- pSB1A3: 3 colonies
- pSB1K3: 60 colonies
- pSB1C3: 9 colonies
- pSB1T3: 0 colonies
- Enhancer (K3): 0 colonies
- GixR (A3): 0 colonies
- Agarose gel
- Agarose gel
- Agarose gel
- Resuspension of a biobrick
- BBa_B0010 (terminator)
- DNA Gel extraction
- CFP
- mRFP
- PCR
- AmiC (primer: MS1 + MS2)
- RBS-Gin (primer: MS12 + MS13)
- Gin (primer: MS11 + MS13)
- ⇒ template-DNA: chromosomal DNA, annealing: 30 sec; 55 °C, elongation: 40 sec; 72 °C, 35 cycles
- Restriction
- pSB1A3 (EcoRI/PstI)
- CFP (EcoRI/PstI)
- mRFP (EcoRI/PstI)
- Transformation
- pSB1A3
- pSB1C3
- pSB1T3
- Terminator
- GixR
- Enhancer
- Inoculation
- pSB1A3
- pSB1C3
- pSB1K3
- Ligation
- pSB1A3 + CFP
- pSB1A3 + mRFP
- GixR Hybridization
- 5 µl primer MS7 (100 µM) + 5 µl primer MS8 (100 µM)
- 5 min 99°C
- over night, room temperature
- -80°C
- DNA Gel extraction
- AmiC
- Ligation
- AmiC + pSV2 (suicide vector)
01.08.2012
- Result
- transformation
- Only BBa_B0010 colonies present
- Plasmid preparation
- pSB1A3 ⇒ only clone 1, 3 Positive
- pSB1C3 ⇒ all negative
- pSB1K3 ⇒ all negative
- Ligation
- GixR hyb. + pJET2_1
- ⇒ over blunt ends
- Restriction
- GixR hyb. (EcoRI/PstI)
- pSB1C3 (EcoRI/PstI)
- Agarose gel
- Agarose gel
- DNA Gel extraction
- RBS-Gin
- Gin
- AmiC
- Test restriction
- pSB1A3 (EcoRI)
- Restriction
- RBS-Gin (EcoRI/PstI)
- Gin (EcoRI/PstI)
- AmiC (EcoRI/PstI)
- pSB1C3 (EcoRI/PstI)
- pSB1K3 (EcoRI/PstI)
- pSB1T3 (EcoRI/PstI)
- Ligation
- cyclisation of pSB1T3
- Fill-in reaction (primer extension)
- Enhancer
- Ligation
- Enhancer and GixR in pSV2
- Enhancer in pSB1K3
- GixR in pSB1C3
- Agarose gel
- pSB1A3 c2
- pSB1C3 c4
- pSB1K3 c3
- Transformation
- pSB1A3 religation
- CFP in A3
- mRFP in A3
- AmiC in pJET
- GixR in pJET
- Enhancer in pJET
- GixR in C3
- Enhancer in K3
- Ligation
- AmiC + pSB1T3
- RBS-Gin + pSB1K3
- Gin + pSB1K3
02.08.2012
- Resuspension of a biobrick
- pSB3C5
- ⇒ for ccdB
- Plasmid preparation
- BBa_B0010 (terminator)
- Test restriction
- Terminator c1,2,3,4 (EcoRI/PstI)
- 2% Agarose gel
- Transformation
- pSB1T3 recycled
- AmiC in T3
- Gin-RBS in K3
- Gin in K3
- pSB3C5 linear (false)
03.08.2012
- Plasmid preparation
- Enhancer in pJET
Week 5
06.08. - 12.08.2012
06.08.2012
- Resuspension and transformation of a biobrick
- BBa_J04450 in pSB1A3
- BBa_J04450 in pSB1C3
- BBa_J04450 in pSB1K3
- BBa_J04450 in pSB1T3
- Restriction
- GFP-fusion (EcoRI/SpeI)
- Terminator (XbaI/PstI)
- Promoter BBa_J23100 (EcoRI/SpeI)
- Promoter BBa_J23108 (EcoRI/SpeI)
- RBS (XbaI/PstI)
- Ligation
- RBS-Gin in T3
- Gin in T3
- GFP-fusion (EcoRI/SpeI) + Terminator (XbaI/PstI) in pSB1K3
- Promoter BBa_J23108 (EcoRI/SpeI) + RBS (XbaI/PstI) in pSB1K3
- Promoter BBa_J23100 (EcoRI/SpeI) + RBS (XbaI/PstI) in pSB1K3
- GixR Hybridization
- 1(3) µl primer MS7 (100 µM) + 1(3) µl primer MS8 (100 µM) + T4-Ligasebuffer
- 5 min 95°C
- over night, room temperature
- -80°C
- Transformation
- GixR in C3
- Gin in T3
- RBS-Gin in T3
- mRFP in A3
- CFP in A3
07.08.2012
- Transformation
- GFP-fusion
- CFP
- J23108-RBS
- J23100-RBS
- Plasmid preparation
- GixR 1,2,3
- Restriction
- GixR hyb. 1,2,3 (EcoRI/PstI)
- pSB1C3 (EcoRI/PstI)
- Ligation
- GixR hyb. 1,2,3 in pJET
- GixR hyb. 1,2,3 in C3
- Transformation
- GixR hyb. 1,2,3 in pJET
- GixR hyb. 1,2,3 in C3
- Test restriction
- pSB3C5 (XbaI)
- pJET (XbaI)
- 1% Agarose gel
- ⇒ positive: pSB3C5 3,4,7,8,; Enhancer-pJET 9-12
08.08.2012
- Result transformation
- GFP-fusion (20 clones)
- J23108-RBS (20 clones)
- J23100-RBS (20 clones)
- GixR hyb. 1,2,3 in pJET (10 clones)
- GixR hyb. 1,2,3 in C3 (10 clones)
- CFP (5 clones)
- Restriktion
- Enhancer in pJET (EcoRI/PstI)
- pSB1C3 (EcoRI/PstI)
- 2% Agarose gel
- DNA Gel extraction
- Enhancer
- pSB1C3
- Ligation
- Enhancer 1,2 in C3
- Transformation
- Enhancer 1,2 in C3
- C3
- mRFP
09.08.2012
- Result transformation
- GFP-fusion ⇒ positive
- J23108-RBS ⇒ positive
- J23100-RBS ⇒ positive
- CFP ⇒ positive
- GixR in C3 c1 ⇒ negative
- GixR in C3 c2 ⇒ negative
- GixR in C3 c3 ⇒ negative
- Plasmid preparation
- GFP-fusion
- J23108-RBS
- J23100-RBS
- CFP
- Test restriction
- GFP-fusion (EcoRI/PstI)
- J23108-RBS (EcoRI)
- J23100-RBS (EcoRI)
- CFP (EcoRI/PstI)
- 1% Agarose gel
- 1% Agarose gel
- 1% Agarose gel
- 1% Agarose gel
- Inoculation
- GixR in C3
- Gin in T3
- Gin-RBS in T3
- pSB1C3
- Enhancer in C3
- mRFP in A3
Week 6
13.08. - 19.08.2012
13.08.12
- PCR
- GroES (primer: MS5 + MS6b)
- HU (primer: MS3 + MS4b)
- ⇒ with and without DMSO, template-DNA: chromosomal DNA, annealing: 30 sec; 60 °C, elongation: 40 sec; 72 °C, 35 cycles
- 1% Agarose gel
- Transformation
- LacIQ (BBa_I14032)
14.08.12
- Restriction
- pSB1K3 (EcoRI/PstI)
- Enhancer 3,4 (EcoRI/ PstI)
- GFP-fusion (EcoRI/SpeI)
- Terminator (XbaI/PstI)
- Test restriction
- RBS-Gin in T3 (EcoRI/ PstI)
- Ligation
- GFP-fusion (EcoRI/SpeI) + Terminator (XbaI/PstI) in pSB1K3 (EcoRI/PstI)
- Promotor J23108 (EcoRI/SpeI) + RBS (XbaI/PstI) in pSB1K3 (EcoRI/PstI)
- Enhancer 3,4 (EcoRI/ PstI) + pSB1K3 (EcoRI/PstI)
- Transformation
- GFP-fusion-Terminator in pSB1K3
- Promotor-RBS in pSB1K3
- Enhancer 3 in pSB1K3
- Enhancer 4 in pSB1K3
- GixR in pJET
15.08.2012
- PCR
- GroES (primer: MS5 + MS6b)
- HU (primer: MS3 + MS4b)
- ⇒ with and without DMSO, template-DNA: chromosomal DNA, annealing: 30 sec; 55 °C, elongation: 30 sec; 72 °C, 35 cycles
- 1% Agarose gel
- DNA Gel extraction
- HU 1,2
- Resuspension of a biobrick
- BBa_J04450 in pSB1A3
- BBa_J04450 in pSB1T3
- BBa_J04450 in pSB1C3
- BBa_J04450 in pSB1K3
- Transformation
- BBa_J04450 in pSB1A3
- BBa_J04450 in pSB1T3
- BBa_J04450 in pSB1C3
- BBa_J04450 in pSB1K3
- Test restriction
- pSB1T3 (EcoRI/PstI and DpnI)
- pSB1K3 (EcoRI/PstI and DpnI)
- pSB1C3 (EcoRI/PstI and DpnI)
- pSB1A3 (EcoRI/PstI and DpnI)
16.08.2012
- PCR
- GroES (primer: MS5 + MS6b)
- ⇒ with and without DMSO, template-DNA: chromosomal DNA, annealing: 30 sec; 55 °C, elongation: 30 sec; 72 °C , 35 cycles
- 1% Agarose gel
- Plasmid preparation
- P-RBS
- Test restriction
- P-RBS (EcoRI/PstI)
- 1% Agarose gel
- Restriction
- Enhancer 3,4 (EcoRI/ PstI)
- HU (EcoRI/ PstI)
- AmiC (EcoRI/ PstI)
- Promoter, GFP (EcoRI/SpeI)
- RBS; Terminator (XbaI/PstI)
- Ligation
- HU, AmiC in pSB1A3
- Enhancer 3,4 in pSB1K3
- P-RBS in pSB1K3
- GFP fusion-Terminator in pSB1K3
- DNA Gel extraction
- GroES 1, P-RBS
- Transformation
- AmiC
- HU
- Enhancer 3,4, 3 alt,4 alt
- Inoculation
- BBa_J04450 in pSB1A3
- BBa_J04450 in pSB1T3
- BBa_J04450 in pSB1C3
- BBa_J04450 in pSB1K3
- AmiC
- P-RBS
17.08.2012
- Restriction
- GroES (EcoRI/SpeI)
- Ligation
- GroES in pSB1C3
- Transformation
- GroES in pSB1C3
- Test restriction
- P-RBS (EcoRI/PstI)
- BBa_J04450 in pSB1A3 (EcoRI/PstI)
- BBa_J04450 in pSB1T3 (EcoRI/PstI)
- BBa_J04450 in pSB1C3 (EcoRI/PstI)
- BBa_J04450 in pSB1K3 (EcoRI/PstI)
- 1% Agarose gel
- Agarose gel
- pipet scheme
- marker|P-RBS 1,2,3,4| P-RBS 1,2,3,4| marker| BBa_J04450 in pSB1K3 1,2,3,4
(bild 22)
- 1% Agarose gel
Week 7
20.08. - 26.08.2012
20.08.2012
- PCR
- Backbone (primer MS24 + MS25)
- ⇒ template-DNA: pSB1A3, pSB1T3, pSB1C3 and pSB1K3, annealing: 30 sec; 55 °C, elongation: 30 sec; 72 °C , 35 cycles
- 1% Agarose gel
- Transformation
- BBa_JH023
- Inoculation
- GroES
- P-RBS
- GFP-T
- Enhancer 3,4
- AmiC
- P-RBS alt
- GFP-T alt
21.08.2012
- Results
- colonies of BBa_JH023
- inoculations except AmiC and P-RBS alt successful
- PCR
- pSB1A3-BBa_J04450 (template DNA)
- pSB1T3-BBa_J04450 (template DNA)
- pSB1C3-BBa_J04450 (template DNA)
- pSB1K3-BBa_J04450 (template DNA)
- used primer --> MS24 + MS25
- initial denaturation 98°C - 5min
- denaturation: 98°C - 1min; annealing: 55°C - 30s; elongation: 72°C - 2.5min -->repeat 35 times
- 72°C - 5min --> 4°C - 8
- miniprep
- GroES, Enh 3/4 (+old), P-RBS (+old), GFP-T (+old)
- Test restriction
- of miniprep
- over night, 37°C
- 1% Agarose gel
- Inoculation
- E.coli BBa_I14032
- over night, 37°C
22.08.2012
- PCR
- CFP (MS16 + MS17)
- mRFP (MS14 + MS15)
- initial denaturation 98°C - 5min
- denaturation: 98°C - 10s; annealing: 60°C - 30s; elongation: 72°C - 40s -->repeat 35 times
- 72°C - 5min --> 4°C - 8
- 1% Agarose gel
- miniprep
- of LacIQ
- Test restriction
- of LacIQ with EcoRI and PstI
- 1% Agarose gel
- 1% Agarose gel
- 1% Agarose gel
- 1% Agarose gel
- test restriction of Enhancer (->21.08.12), gel 2%
- watch picture
- test restriction of Enhancer and P-RBS (->21.08.12), gel 2%
- watch picture
23.08.2012
- PCR
- pSB1K3-BBa_J04450 (template DNA)
- pSB1T3-BBa_J04450 (template DNA)
- used primer --> MS24 + MS25
- initial denaturation 98°C - 5min
- denaturation: 98°C - 1min; annealing: 55°C - 30s; elongation: 72°C - 2.5min -->repeat 35 times
- 72°C - 5min --> 4°C - 8
- 1% Agarose gel
- Restriction
- GixR (EcoRI/PstI)
- Ligation
- GixR in pSB1C3
- Transformation
- GixR in pSB1C3
24.08.2012
- PCR
- pSB1A3-BBa_J04450 (template DNA)
- pSB1T3-BBa_J04450 (template DNA)
- pSB1C3-BBa_J04450 (template DNA)
- pSB1K3-BBa_J04450 (template DNA)
- initial denaturation 98°C - 5min
- denaturation: 98°C - 10s; annealing: 60°C - 30s; elongation: 72°C - 40s -->repeat 30 times
- 72°C - 10min --> 4°C - 8
- primer: MS22 + MS23
- 1% Agarose gel
- Restriction
- RBS (XbaI/PstI)
- CFP (EcoRI/PstI)
- mRFP (EcoRI/PstI)
- Ligation
- mRFP und CFP in pSB1C3
- Transformation
- mRFP und CFP in pSB1C3
Week 8
27.08. - 02.09.2012
27.08.2012
- Results
- CFP: 1 colony
- CFP (neu): Colonies
- mRFP: colonies
- mRFP (neu): no colonies
- PCR
- template: backbones (pSB1K3/T3/C3/A3)
- parameters: 98°C 5min, >> 98°C 1min, 70°C 30s, 72°C 2.5min << x30, 72°C 5min, 4°C 8
- primer: MS22 + MS23
- colony-PCR
- parameters: 98°C 5min, >> 98°C 1min, 72°C 1min << x30, 72°C 5min, 4°C 8
- primer: MS18 + MS19
- Agarose gel of backbone PCR
- no result
- elution of the backbones of Friday the 24th
- 1% Agarose gel
- 1% Agarose gel
- Inoculation
- pSB1C3_...
- mRFP clone 20,24
- CFP clone 14,15,17,18
- gix clone 1,2
- 5ml LB, over night, 37°C
28.08.2012
- production of competent TOP10
- PCR
- Enhancer, Terminator, Gix, P-RBS
- parameters: 98°C 5', 98°C 1', 66°C 30", 72°C 30", 72°C 5', 4°C ~
- primer: MS20 + MS21
- 1% Agarose gele
- Transformation
- pBad, araC, pSacB
- over night, 37°C
- miniprep
- GixR (1,2)
- CFP (14,15,17,18)
- mRFP (20,24)
- alle in pSB1C3
29.08.2012
- Results
- Transformation: no colonies
- PCR
- Terminator, Gix, Enhancer, P-RBS
- diluted templates: Terminator, P-RBS, Gix 1 --> 1:10
- Enhancer, Gix 2 --> 1:20
- parameters: 98°C 5', 98°C 1', 66°C 30", 72°C 30", 72°C 5', 4°C ~
- primer: MS20 + MS21
- 1% Agarose gele
- 3AA
- CFP, mRFP with Terminator
- 2µl DNA
- 1µl SpeI
- 2µl Tango
- 15µl dH2O
- --> 1h at 37°C
- +1µl EcoRI
- +2,5µl Tange
- --> 1h at 37°C
- --> inactivation for 20min at 80°C
- Ligation
- 3AA products in pSB1T3
- 18.5µl H2O
- 2.5 µl Buffer
- 1µl Insert CFP or mRFP (EcoRI + SpeI)
- 1µl Insert Terminator (BBa_B0010) (XbaI + PstI)
- 1µl pSB1T3 (EcoRI + PstI)
- 1µl Ligase
- --> 1h at RT
- Transformation
- pBad --> TOP10 + pSB2K3
- CFP-T, mRFP-T --> TOP10 + pSB1T3
- Sequencing
- CFP_fwd
- CFP_rev
- mRFP_fwd
- mRFP_rev
- P-RBS_fwd
- GixR_fwd
- GixR_fwd
- Enhancer_fwd
30.08.2012
- Results
- Transformation: no colonies --> testing Trafo with BBa_J04450 A/K/T/C
- 2 colonies at pBad-Trafo from 28.08.2012 --> liquid culture for miniprep
- 1 colony at pSacB-Trafo from 28.08.2012 --> tested on succrose-sensitivity
31.08.2012
- PCR
- SacB
- Parameters No.1: 95°C 5', >>95°C 30", 60°C 30", 72°C 1'<< x30, 72°C 5', 4°C
- Parameters No.2: 95°C 5', >>95°C 30", 72°C 1.5'<< x30, 72°C 5', 4°C
- Primer MS26 + MS27
- 1% Agarose gele
- Ligation
- 3AA (CFP-T, mRFP-T)
- watch 29.08.
- used vector: pSB1K3/T3
- Transformation
- of ligation in TOP10
- miniprep
- pBad/araC K.1+2
- restriction
- GixR rehybridization
- cut with EcoRI and PstI
- ligation
- 1µl GixRhyb E+P
- 1µl pSB1C3 E+P
- 1µl Th-Pol
- 2µl Buffer
- 15µl dH2O
- Transformation
- ligation + TOP10 --> LB(cam)
- Results of test-trafo
- Amp, Km, Cm OK --> Km with fewer colonies
- Tet --> hardly colonies
- --> comp.TOP10 obviously all right, exclusion of Tet in future work
Week 9
03.09. - 09.09.2012
03.09.2012
- Results
- Transformation
- GixR in C3 --> no colonies
- CFP-T/mRFP-T in C3(31.8.) --> no colonies
- CFP-T/mRFP-T in T3(31.8.) --> many but small colonies
- CFP-T/mRFP-T in T3(29.8.) --> less colonies
- colony-PCR
- of colonies (CFP-T, mRFP-T)
- parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C
- primer MS18 + MS19
- Agarose gel
- 6x colony pT3-CFP-T from 29./31.8.
- 6x colony pT3-mRFP-T from 29./31.8.
- pC3-CFP (14)
- pC3.mRFP (24)
- 1% Agarose gele
Week 10
10.09. - 16.09.2012
11.09.2012
- microscopy-screening
- GroES-GFP ⇒; no fluorescence
- GroES-CFP ⇒; no fluorescence
- AmiC-mRFP ⇒; no fluorescence
- AmiC-GFP ⇒; no fluorescence
- colony-PCR
- GixR
- RBS-Gin
- Terminator
- parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C
- primer MS20 + MS21
- 3A-Assembly (digestion, ligation, transformation)
- GroES (E/S) + Terminator (X/P) + pSB1K3 (P/E)
- AmiC (E/S) + Terminator (X/P) + pSB1K3 (P/E)
- plasmid-prep and control-digestion (E/P)
- GroES-GFP
- GroES-CFP
- AmiC-mRFP
- AmiC-GFP
- GroES-Terminator
- Enhancer
- GFP-Terminator
- Terminator
12.09.2012
- Results
- Transformation
- - P-RBS --> colonies
- - pB10 --> no colonies
- - RBS (B0030) --> colonies
- - Gro-ES/ AmiC-Terminator --> no colonies
- colony-PCR
- Gix (2. try)
- Gin-RBS (2. try)
- RBS (B0030)
- P-RBS (108)
- parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C
- primer MS20 + MS21
- Agarose gel
- Gin-RBS (5x)
- Gin (1x)
- Gix (5x)
- RBS (6x)
- P-RBS (6x)
- 1% Agarose gele
- 1% Agarose gele
- Restriction (miniprep 11.09.12)
- gr-GFP
- gr-CFP
- gr-T K3
- enhancer K3
- am-RFP-K3
- am-GFP-K3
- GFP-T-C3
- terminator A2
- 1% Agarose gele
- 1% Agarose gele
- Transformation
- groES-T
- 2 Transformations:
- - One with a 1:10 dilution of Terminator from ligation,
- - another one with a 1:100 dilution of Terminator from ligation
- 2 Transformations:
- amiC-T
- 2 Transformations:
- - One with a 1:10 dilution of Terminator from ligation,
- - another one with a 1:100 dilution of Terminator from ligation
- 2 Transformations:
- PCR
- Gin-RBS
- 3 reactions:
- -One using High Fidelity (HF) Reaction Buffer,
- -one using GC-Buffer
- -and another one using self-made reaction buffer
- 3 reactions:
- Primer used : MS12 + MS13
- Template DNA : E. coli C600 chromosomal DNA
13.09.2012
- Results
- Transformation
- - pBAD (11.09.12) --> no colonies
- - AmiC-T (11.09.12) --> 2 x 2 colonies
- - GroES-T (11.12.09)--> 2 colonies
- colony-PCR
- AmiC-T & GroES-T (transformation 11.09.12)
- parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C
- primer MS20 + MS21
- 1% Agarose gele
- Gel extraction
- RBS-Gin (HF) --> 51 ng/µl
- RBS-Gin (GC) --> 69 ng/µl
- RBS-Gin (SM) --> 56 ng/µl
- Ligation
- GroES-T & AmiC-T in pSB1K3
- Restriction (preparative digest)
- GroES (A module)
- with SpeI
- AmiC (A module)
- with SpeI
- mRFP-C3 (B module)
- with EcoRI and XbaI
- CFP-C3 (B module)
- with EcoRI and XbaI
- Gin-RBS
- HF-PCR-Product, GC-PCR-product and SM-PCR-product
- with EcoRI and PstI
- HF-PCR-Product, GC-PCR-product and SM-PCR-product
- 1% Agarose gele
14.09.2012
- Results
- Transformation
- AmiC-T: no colonies
- GroES-T: no colonies
- Restriction
- 2nd Digestion of A-modules (see digest from yesterday) additionally with EcoRI
- Ligation
- Gin-RBS with pSB1C3
- 3 reactions:
- one with RBS-Gin HF-PCR-Product
- one with RBS-Gin GC-PCR-Product
- one with RBS-Gin SM-PCR-Product
- 3 reactions:
- Inoculation
- Gix-K3 (clone 3 and 4)
- P-RBS-C3 (clone 1 and 3)
- RBS (BBa_B0030) [clone 1 and 3]
- 1% Agarose gele
- Gel extraction
- GFP
- mRFP
- 1% Agarose gele
- Gel extraction
- Terminator clone 48
- Transformation
- RBS-Gin(HF)-pSB1C3
- RBS-Gin(GC)-pSB1C3
- RBS-Gin(SM)-pSB1C3
Week 11
17.09. - 23.09.2012
17.09.2012
- Restriction
- AmiC
- GroES
- CFP
- SacB
- 1% Agarose gele
- Restriction
- CFP-C3
- SacB (2nd try)
- Resuspend
- BBa_B001J
- Restreak
- RBS-Gin (HF)
- RBS-Gin (GC)
- RBS-Gin (SM)
- DNA isolation
- CFP-C3 (Miniprep28.08.12)--> 47,3 ng/µl
- Miniprep
- GixR pSB1K3 --> 50,5 ng/µl, 144,7 ng/µl
- RBS(new)B0030 pSB1A2 --> 726,2 ng/µl, 242,6 ng/µl
- P108-RBS pSB1C3 --> 200,5 ng/µl, 177,8 ng/µl
- 1% Agarose gele
- Transformation
- pSB2K3-pBAD & pSB1-A2-TT ???????? kann ich nicht lesen
- 1% Agarose gel
- CFP-C3 (restriction 2nd try)
- SacB-Cs (restriction 2nd try)
18.09.2012
- Results
- Transformation
- 2x Terminator --> colonies
- pBAD --> no colonies
- Colony PCR
- RBS-Gin
- 2x Terminator
- parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C
- primer MS20 + MS21
- 1% Agarose gele
- Sequencing
- SacB
- Gel extraction
- CFP-C3 E+X
- Ligation
- am-G: amiC E+S & GFP-A3 E+X
- am-C: amiC E+S & CFP-C3 E+X
- am-R: amiC E+S & mRFP-C3 E+X
- gro-G: groES E+S & GFP-A3 E+X
- gro-C: groES E+S & CFP-C3 E+X
- gro-R: groES E+S & mRFP-C3 E+X
- Transformation
- am-G
- am-C
- am-R
- gr-G
- gr-C
- gr-R
- pBAD
19.09.2012
- Results
- Transformation
- am-G --> colonies
- am-C --> colonies
- am-R --> colonies
- gr-G --> colonies
- gr-C --> colonies
- gr-R --> colonies
- pBAD --> no colonies
- Colony PCR
- RBS-Gin (from 17.09.12)
- 2x Terminator B0017 (from 17.09.12)
- parameters: 98°C 5', >>98°C 1', 72°C 1'<<x30, 72°C 5', 4°C
- primer MS20 + MS21
- --> Amplification failed
- Transformation
- pBAD
- Microscopy-screening
- AmiC-GFP --> fluorescence
- GroES-GFP --> fluorescence
- AmiC-CFP --> fluorescence
- GroES-CFP --> fluorescence
- AmiC-mRFP --> fluorescence
- GroES-mRFP --> fluorescence
- Inoculation
- RBS-Gin
- TT (B0017)
- AmiC-GFP
- GroES-GFP
- AmiC-CFP
- GroES-CFP
- AmiC-mRFP
- GroES-mRFP
20.09.2012
- Miniprep
- RBS-Gin
- TT (B0017)
- AmiC-GFP
- GroES-GFP
- AmiC-CFP
- GroES-CFP
- AmiC-mRFP
- GroES-mRFP
- Restriction (E/P)
- TT(B0017)
- RBS-Gin
- 1% Agarose gele
- Restriction
- SacB ((XbaI + PstI) & (EcoRI + XbaI)
- P-RBS (EcoRI + SpeI) & (SpeI + PstI)
- 1% Agarose gel
- Inoculation
- GroES-GFP
- GroES-CFP
- GroES-mRFP
21.09.2012
- Restriction
- GFP-fusion (EcoRI + SpeI)
- TT BBa_0017 (EcoRI + XbaI)
- 1% Agarose gel
- Gel extraction
- GFP-Fusion
- pSB1A2 (backbone GFP-Fusion)
- TT
- SacB
- Ligation
- GFP bb Enhancer
- GFP TT
- GroES TT
- Transformation
- GFP bb Enhancer
- GFP TT
- GroES TT
- Promotor-RBS (17.09.12)
- Sequencing
- RBS-Gin
- Microscopy-screening
- AmiC-GFP --> Channel: DIC, Green, Red, Blue
- GroES-GFP --> Channel: DIC, Green, Red, Blue
- AmiC-CFP --> Channel: DIC, Green, Red, Blue
- GroES-CFP --> Channel: DIC, Green, Red, Blue
- AmiC-mRFP --> Channel: DIC, Red
- GroES-mRFP --> Channel: DIC, Green, Red, Blue
22.09.2012
- Inoculation
- GroES-TT pSB1A2
- GFP-TT pSB1A2
- Enhancer
- P108-RBS pSB1C3
 "
"