Team:ZJU-China/project.htm
From 2012.igem.org
(Difference between revisions)
| Line 425: | Line 425: | ||
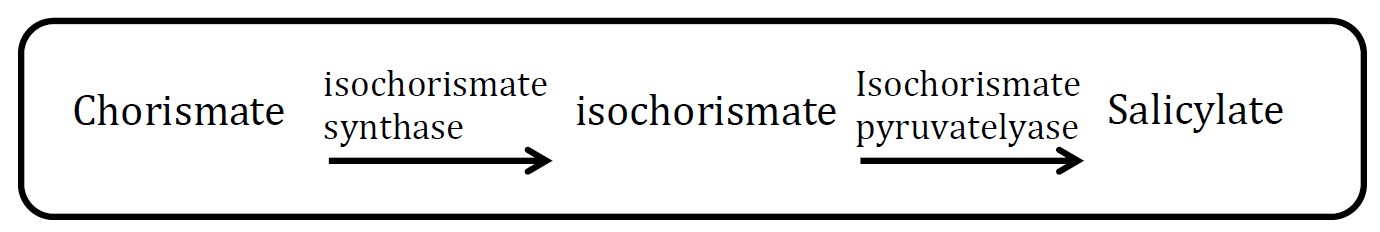
<h2>1. Salicylate pathway</h2> | <h2>1. Salicylate pathway</h2> | ||
<h2>(Group: iGEM2006_MIT)</h2> | <h2>(Group: iGEM2006_MIT)</h2> | ||
| + | <img src="http://www.jiajunlu.com/igem/zju_iaa_1.jpg" width="600px" /> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">Assessment: </p> | <p align="justify">Assessment: </p> | ||
| Line 432: | Line 433: | ||
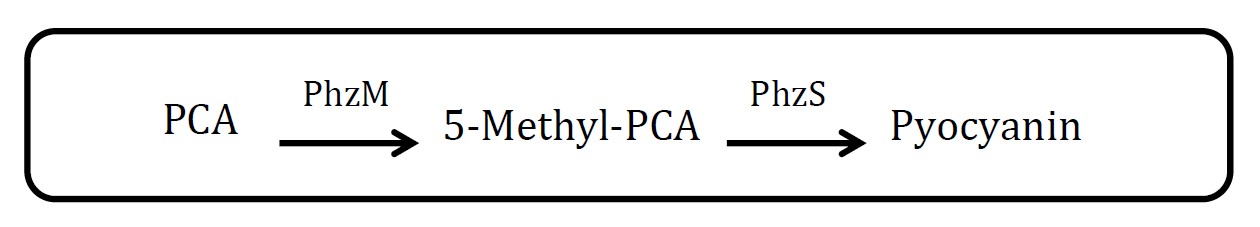
<h2>2. Pyocyanin pathway</h2> | <h2>2. Pyocyanin pathway</h2> | ||
<h2>(Group: iGEM2007_Glasgow)</h2> | <h2>(Group: iGEM2007_Glasgow)</h2> | ||
| + | <img src="http://www.jiajunlu.com/igem/zju_iaa_2.jpg" width="600px" /> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">Assessment: </p> | <p align="justify">Assessment: </p> | ||
| Line 439: | Line 441: | ||
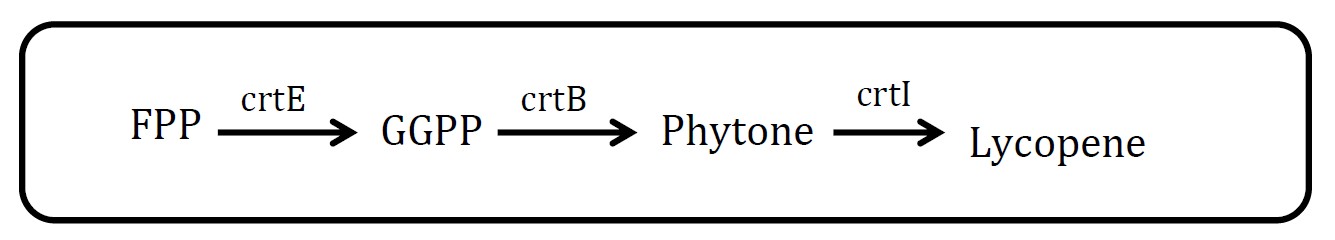
<h2>3. Lycopene pathway</h2> | <h2>3. Lycopene pathway</h2> | ||
<h2>(Group: iGEM2009_Cambridge) </h2> | <h2>(Group: iGEM2009_Cambridge) </h2> | ||
| + | <img src="http://www.jiajunlu.com/igem/zju_iaa_3.jpg" width="600px" /> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">Assessment: </p> | <p align="justify">Assessment: </p> | ||
| Line 444: | Line 447: | ||
<p align="justify">Lycopene is visible red and its substrate, FPP, is colorless. So measurement is quite feasible. But there are at least three proteins in this pathway, which will increase the burden of cell. But in future work, we could have a try.</p> | <p align="justify">Lycopene is visible red and its substrate, FPP, is colorless. So measurement is quite feasible. But there are at least three proteins in this pathway, which will increase the burden of cell. But in future work, we could have a try.</p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
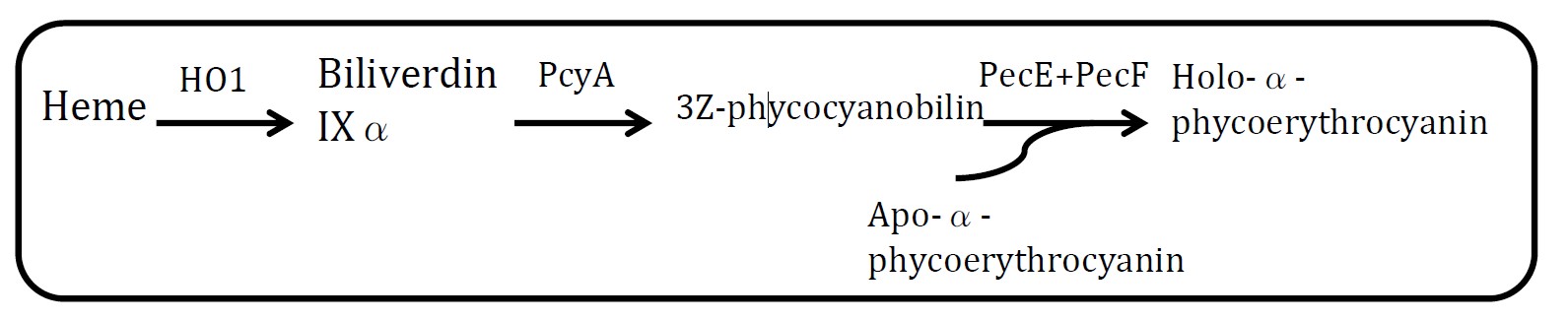
| - | <h2>4. Holo- | + | <h2>4. Holo-α-phycoerythrocyanin pathway</h2> |
<h2>(Group: iGEM2004_UTAustin)</h2> | <h2>(Group: iGEM2004_UTAustin)</h2> | ||
| + | <img src="http://www.jiajunlu.com/igem/zju_iaa_4.jpg" width="600px" /> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">Assessment: </p> | <p align="justify">Assessment: </p> | ||
| Line 460: | Line 464: | ||
<h2>6. IAM pathway</h2> | <h2>6. IAM pathway</h2> | ||
<h2>(Group: iGEM2011_Imperial)</h2> | <h2>(Group: iGEM2011_Imperial)</h2> | ||
| + | <img src="http://www.jiajunlu.com/igem/zju_iaa_5.jpg" width="600px" /> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
<p align="justify">Assessment: </p> | <p align="justify">Assessment: </p> | ||
<p align="justify"> </p> | <p align="justify"> </p> | ||
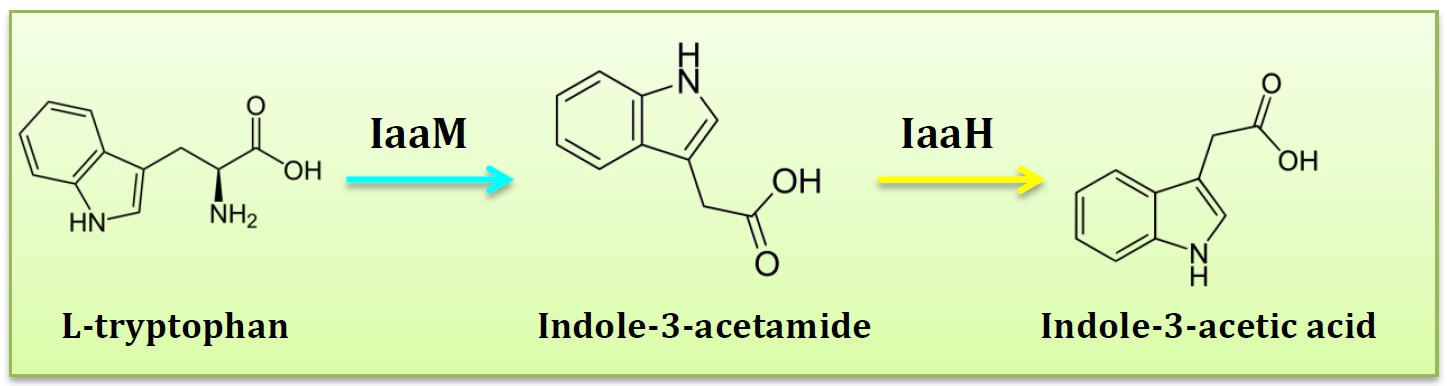
<p align="justify">Five pathways described above all have some drawbacks, finally, only one pathway left, IAM pathway. The two-step IAM pathway generates indole-3-acetic acid (IAA) from the precursor tryptophan. IAA tryptophan monooxygenase (IaaM) catalyses the oxidative carboxylation of L-tryptophan to indole-3-acetamide, which is hydrolysed to IAA and ammonia by indoleacetamide hydrolase (IaaH). </p> | <p align="justify">Five pathways described above all have some drawbacks, finally, only one pathway left, IAM pathway. The two-step IAM pathway generates indole-3-acetic acid (IAA) from the precursor tryptophan. IAA tryptophan monooxygenase (IaaM) catalyses the oxidative carboxylation of L-tryptophan to indole-3-acetamide, which is hydrolysed to IAA and ammonia by indoleacetamide hydrolase (IaaH). </p> | ||
| + | <p align="justify"> </p> | ||
| + | <p align="justify">Final Decision: </p> | ||
| + | <p align="justify"> </p> | ||
| + | <img src="http://www.jiajunlu.com/igem/zju_iaa_6.jpg" width="600px" /> | ||
</div> | </div> | ||
</div><!-- end .acc_container --> | </div><!-- end .acc_container --> | ||
Revision as of 14:08, 26 September 2012

 "
"