Team:LMU-Munich/Inverter
From 2012.igem.org
| Line 10: | Line 10: | ||
The LMU-Munich 2011 Team started a project to convert a positive input signal into a negative output or a negative input signal into a positive output. | The LMU-Munich 2011 Team started a project to convert a positive input signal into a negative output or a negative input signal into a positive output. | ||
| - | Last year LMU-Munich designed a metal-sensing device. However, it turned out that some of the promoters that respond to metal ions are | + | Last year the LMU-Munich iGEM team designed a metal-sensing device. This device links metal sensing promoters to a visual output. However, it turned out that some of the promoters that respond to metal ions are negatively regulated. So there was a need of an inverter, a genetic part that converts the repression of the metal sensing promoter into a positive output. |
Since such a genetic part would be a very beneficial tool, Julia, who also participated in iGEM 2011, continued the work from last year and got some great results. | Since such a genetic part would be a very beneficial tool, Julia, who also participated in iGEM 2011, continued the work from last year and got some great results. | ||
| - | * To get an explanation how it works and see the results: [[Team:LMU-Munich/Inverter#Theory and Results|Data and Results]] | + | * To get an explanation of how it works and see the results: [[Team:LMU-Munich/Inverter#Theory and Results|Data and Results]] |
| - | * How to compose your individual Inverter, | + | * How to compose your individual Inverter, by fusions PCRs and 3a assemblies is explained here: [[Team:LMU-Munich/Inverter#Construct your own Inverter|Construct your own Inverter]] |
| Line 21: | Line 21: | ||
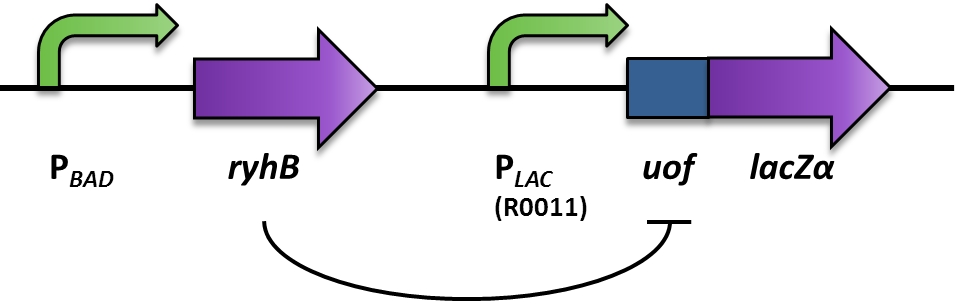
[[File:LMU Inverter Construct.png|600px]] | [[File:LMU Inverter Construct.png|600px]] | ||
| - | The [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] is | + | The main component of the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] is the small RNA RyhB which translationally inhibits the upstream fused region ''uof<sub>CGU</sub>'' by binding to it and therefore masking Shine Dalgarno sequence. The promoter one wants to invert, in this proof of principle case it is the arabionose-inducible P<sub>''BAD''</sub> (PCR of [http://partsregistry.org/Part:BBa_I0500 BBa_I0500]), regulates the expression of the small RNA RyhB (PCR of pURyhB) ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1852835/ Vecerek B., Bläsi U., 2007]). RyhB itself translationally inhibits uof<sub>CGU</sub> (upstream of ''fur'') (PCR of pRuof<sub>CGU</sub>-lacZ) ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1852835/ Vecerek B., Bläsi U., 2007]) by binding and masking the Shine Dalgarno sequence. If uof<sub>CGU</sub> is translationally fused to a reporter, in this case ''lacZalpha'' (PCR from pRuof<sub>CGU</sub>-''lacZ'' from [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1852835/ Vecerek B., Bläsi U., 2007]), expression of the beta-Galactosidase is translationally repressed. The repression of the reporter is dependent on the concentration of RyhB, and therefore the induction of the P<sub>''BAD''</sub> promoter. consequently, the induction of the P<sub>''BAD''</sub> promoter is inverted to a negative output of the reporter. |
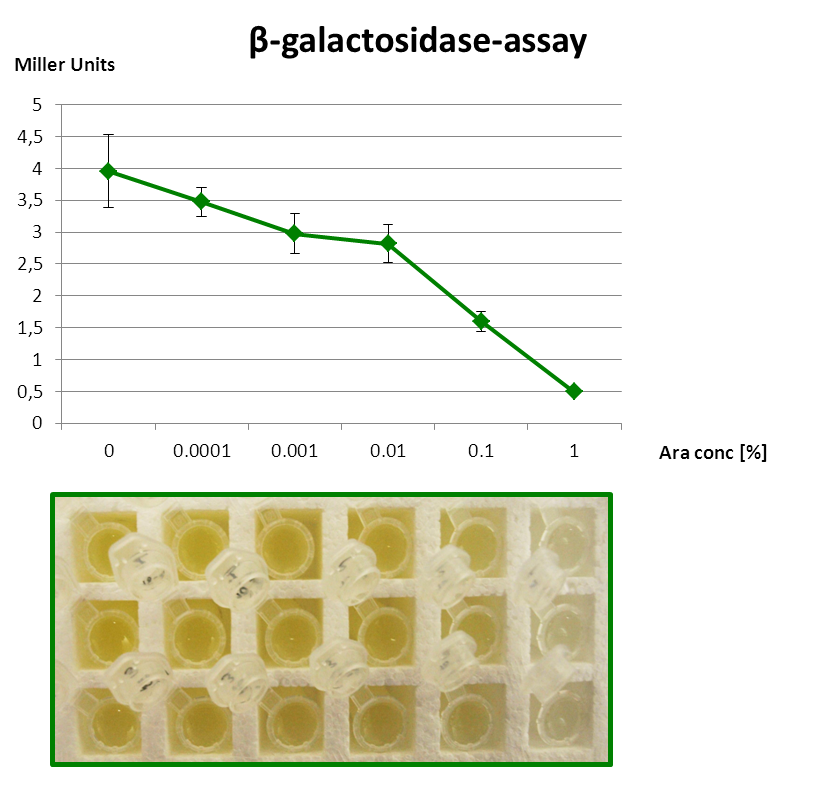
| - | The | + | The Beta-galatosidase assay below shows the function of this Inverter. In all cases grown with 1 mM IPTG so always the Reporter is induced fully. When grown with Arabinose in the media, RyhB is produced and this inhibits uof<sub>CGU</sub> and consequently the fused reporter. The more Arabinose the more the translational repression. |
| + | |||
| + | But as the P<sub>''BAD</sub> promoter lacks the repressor binding sites, it is leaky and always produces a bit RyhB and therefore there is always a repression of the reporter. Consequently we get only a small signal without Arabinose (minimal repression) and the P<sub>BAD</sub> promoter is generally poorly titratable. But as this peticuliar Inverter is just a proof of principle, it is not essential to be such. The Inverter for the wanted Promoter and Output has to be constructed by fusion PCR (see next section). | ||
[[File:LMU Inverter graph.png|620px]] | [[File:LMU Inverter graph.png|620px]] | ||
| Line 39: | Line 41: | ||
* basic PCRs: Promoter (a+b), RyhB (c+d), uof (e+f), Output/Reporter (g+h) | * basic PCRs: Promoter (a+b), RyhB (c+d), uof (e+f), Output/Reporter (g+h) | ||
| - | * Fusion PCRs: ~ 200 ng equimolar with forward primer of the front and reverse primer of the back fusion part: | + | * Fusion PCRs: ~ 200 ng equimolar with forward primer of the front and reverse primer of the back fusion part: promoter to RyhB (a+d) and uof to Output/Reporter (e+h) |
3. Bring these into BioBrick vectors to facilitate 3a assemblies | 3. Bring these into BioBrick vectors to facilitate 3a assemblies | ||
Revision as of 12:19, 26 September 2012
The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


Inverter
The LMU-Munich 2011 Team started a project to convert a positive input signal into a negative output or a negative input signal into a positive output.
Last year the LMU-Munich iGEM team designed a metal-sensing device. This device links metal sensing promoters to a visual output. However, it turned out that some of the promoters that respond to metal ions are negatively regulated. So there was a need of an inverter, a genetic part that converts the repression of the metal sensing promoter into a positive output.
Since such a genetic part would be a very beneficial tool, Julia, who also participated in iGEM 2011, continued the work from last year and got some great results.
- To get an explanation of how it works and see the results: Data and Results
- How to compose your individual Inverter, by fusions PCRs and 3a assemblies is explained here: Construct your own Inverter
Theory and Results
The main component of the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823040 Inverter] is the small RNA RyhB which translationally inhibits the upstream fused region uofCGU by binding to it and therefore masking Shine Dalgarno sequence. The promoter one wants to invert, in this proof of principle case it is the arabionose-inducible PBAD (PCR of [http://partsregistry.org/Part:BBa_I0500 BBa_I0500]), regulates the expression of the small RNA RyhB (PCR of pURyhB) ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1852835/ Vecerek B., Bläsi U., 2007]). RyhB itself translationally inhibits uofCGU (upstream of fur) (PCR of pRuofCGU-lacZ) ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1852835/ Vecerek B., Bläsi U., 2007]) by binding and masking the Shine Dalgarno sequence. If uofCGU is translationally fused to a reporter, in this case lacZalpha (PCR from pRuofCGU-lacZ from [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1852835/ Vecerek B., Bläsi U., 2007]), expression of the beta-Galactosidase is translationally repressed. The repression of the reporter is dependent on the concentration of RyhB, and therefore the induction of the PBAD promoter. consequently, the induction of the PBAD promoter is inverted to a negative output of the reporter.
The Beta-galatosidase assay below shows the function of this Inverter. In all cases grown with 1 mM IPTG so always the Reporter is induced fully. When grown with Arabinose in the media, RyhB is produced and this inhibits uofCGU and consequently the fused reporter. The more Arabinose the more the translational repression.
But as the PBAD promoter lacks the repressor binding sites, it is leaky and always produces a bit RyhB and therefore there is always a repression of the reporter. Consequently we get only a small signal without Arabinose (minimal repression) and the PBAD promoter is generally poorly titratable. But as this peticuliar Inverter is just a proof of principle, it is not essential to be such. The Inverter for the wanted Promoter and Output has to be constructed by fusion PCR (see next section).
Construct your own Inverter
1. Primer design:
- Promoter (which you would like to invert): BB-suffix + Promoter primer forward (a), GTCTTCCTGATCGCGAGACA + Promoter reverse primer from -10 (b)
- RyhB: TGTCTCGCGATCAGGAAGAC (RyhB fwd) (c), BB-suffix + AAAGCCAGCACCCGGCTGGCTAAG (RyhB rev) (d)
- uof: BB-prefix + GTGGTTTTCATTTAGGCGTG (Uof fwd) (e), Output/Reporter forward primer + GTTATCAGTCATGCGGAATC (uof rev) (f)
- Output/Reporter: Forward primer without BB-prefix (g), Reverse primer with BB-suffix (h)
2. PCRs and Fusion PCRs:
- basic PCRs: Promoter (a+b), RyhB (c+d), uof (e+f), Output/Reporter (g+h)
- Fusion PCRs: ~ 200 ng equimolar with forward primer of the front and reverse primer of the back fusion part: promoter to RyhB (a+d) and uof to Output/Reporter (e+h)
3. Bring these into BioBrick vectors to facilitate 3a assemblies
4. 3a assemblies
- 3a assembly of Output/Reporter with a constitutive/inducible promoter like [http://partsregistry.org/Part:BBa_R0011 BBa_R0011]
- 3a assembly of promoter + RyhB with product of step above
 "
"