Team:LMU-Munich/Bacillus BioBricks
From 2012.igem.org
| Line 208: | Line 208: | ||
<br> | <br> | ||
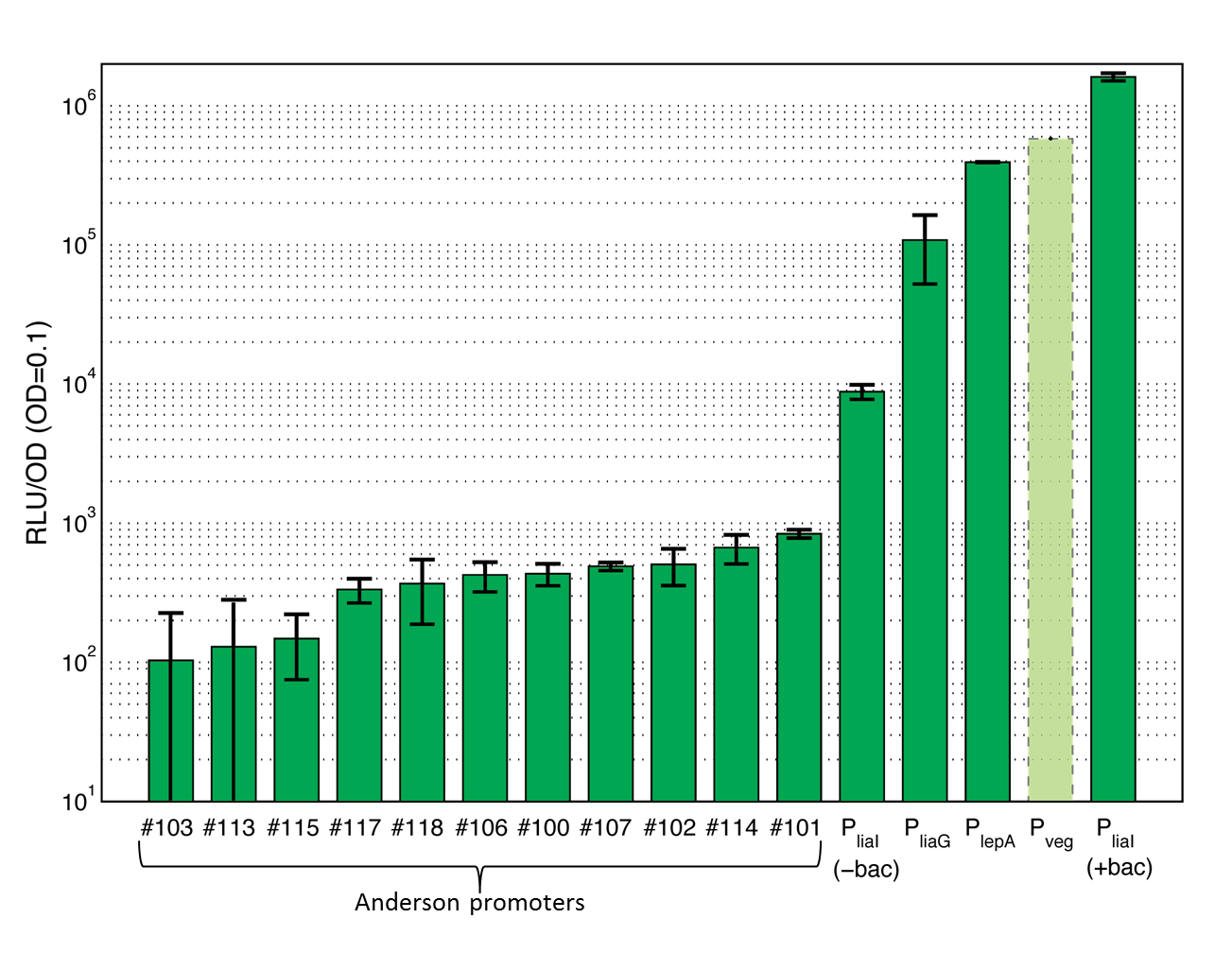
| - | [[File:Promoters overview.png| thumb|center|600px|<p align="justify"> '''Overview of promoter activity measured with luminescence measurements.''' These values derive from the experiments you can find in our Data section. Lumi per OD<sub>''600''</sub> are taken at a OD<sub>600</sub> of 0,1. Curves are the average and the standard deviation of two independent clones. Shown is the activity of the Anderson promoters J23100 (#100), J23101 (#101), J23102 (#102), J23103 (#103), J23106 (#106), J23107 (#107), J23113 (#113), J23114 (#114), J23115 (#115), J23117 (#117), J23118 (#118) as well as the activity of the constitutive promoters P<sub>''liaG''</sub>, and P<sub>''lepA''</sub>. The activity of the inducible promoter P<sub>''liaI''</sub> is shown with (+bac) and without (-bac) induction with bacitracin (10 μg/ml). The promoter activity of P<sub>''veg''</sub> is projected from the results from the beta-galactosidase assay and was not measured with luminescence measurements. </p> | + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" |
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[File:Promoters overview.png|thumb|center|600px]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="70%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
| + | <font color="#000000"; size="2"><p align="justify"> '''Overview of promoter activity measured with luminescence measurements.''' These values derive from the experiments you can find in our Data section. Lumi per OD<sub>''600''</sub> are taken at a OD<sub>600</sub> of 0,1. Curves are the average and the standard deviation of two independent clones. Shown is the activity of the Anderson promoters J23100 (#100), J23101 (#101), J23102 (#102), J23103 (#103), J23106 (#106), J23107 (#107), J23113 (#113), J23114 (#114), J23115 (#115), J23117 (#117), J23118 (#118) as well as the activity of the constitutive promoters P<sub>''liaG''</sub>, and P<sub>''lepA''</sub>. The activity of the inducible promoter P<sub>''liaI''</sub> is shown with (+bac) and without (-bac) induction with bacitracin (10 μg/ml). The promoter activity of P<sub>''veg''</sub> is projected from the results from the beta-galactosidase assay and was not measured with luminescence measurements. </p></font> | ||
| + | |} | ||
====Anderson promoters==== | ====Anderson promoters==== | ||
Revision as of 13:48, 25 September 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


B4 - 22 core parts for Bacillus subtilis
We will create a toolbox of Bacillus BioBricks to contribute to the registry.
This Bacillus BioBrickBox (B4) contains Bacillus specific parts:
| Vectors | Promoters | Reporters | Affinity tags |

| 
| 
| |
|
pSBBs1C |
Anderson |
gfp |
Flag |
Bacillus Vectors 
We have generated a suit of BioBrick-compatible vectors, three empty ones with different antibiotic resistances and integration loci, two reporter and two expression vectors.Here is a list of all the vectors we cloned and used.
For the use of our vectors, please see our Protocols page. A general introduction to Bacillus subtilis and its integrative vectors can be found here. All vectors have ampicillin as Escherichia coli resistance and RFP in the multiple cloning site as selection marker.
| Vector Name | Resistance | Insertion | Description | Vector origin | ||
|---|---|---|---|---|---|---|
| BioBrick | Eco | Bsu | locus | Name | Reference | |
| pSBBs1C [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823023 (BBa_K823023)] | Amp | Cm | amyE | empty | pDG1662 | [http://www.ncbi.nlm.nih.gov/pubmed/8973347 Guérout-Fleury] |
| pSBBs4S [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823022 (BBa_K823022)] | Amp | Spec | thrC | empty | pDG1731 | [http://www.ncbi.nlm.nih.gov/pubmed/8973347 Guérout-Fleury] |
| pSBBs2E [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823027 (BBa_K823027)] | Amp | MLS | lacA | empty | pAX01 | [http://www.ncbi.nlm.nih.gov/pubmed/11274134 Härtl] |
| pSBBs1C-lacZ [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823021 (BBa_K823021) ] | Amp | Cm | amyE | lacZ reporter | pAC6 | [http://www.ncbi.nlm.nih.gov/pubmed/11902727 Stülke] |
| pSBBs3C-luxABCDE [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823025 (BBa_K823025)] | Amp | Cm | sacA | luxABCDE reporter | pAH328 | [http://www.ncbi.nlm.nih.gov/pubmed/20709900 Schmalisch] |
| pSBBs4S-PXyl [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823024 (BBa_K823024)] | Amp | Spec | thrC | Xylose-promoter | pXT | [http://www.ncbi.nlm.nih.gov/pubmed/11069659 Derré] |
| pSBBs0K-Pspac [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823026 (BBa_K823026)] | Amp | Kan | replicative | IPTG-promoter | pDG148 | [http://www.ncbi.nlm.nih.gov/pubmed/11728721 Joseph] |
| Sporovector [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823054 (BBa_K823054)] | Amp | Spec | thrC | to create Sporobeads | pSBBs4S | Sporovector |
Here you can find the respective vector maps:

| 
|

| 
|

| 
|

| 
|
The number in the vector's name codes for the insertion locus and the following letter for the Bacillus subtilis resistance gene according to the following table:
| Number | Insertion locus | Letter | Resistance |
|---|---|---|---|
| 0 | replicative | C | Chloramphenicol (Cm) |
| 1 | amyE (amylase) | E | MLS (Erythromycin + Lincomycin) |
| 2 | lacA (β-galactosidase) | K | Kanamycin (Kan) |
| 3 | sacA (sucrase) | S | Spectinomycin (Spec) |
| 4 | thrC (threonine synthase) |
The concentrations of the antibiotics and the insertion tests can be found in our Protocol section.
See here to find out how to use B. subtilis vectors. In this overview, the mechanism of integration B. subtilis vectors is described.
The design and special use of our Sporovector can be found here.
For results, check our data page: ![]()
Bacillus Promoters 
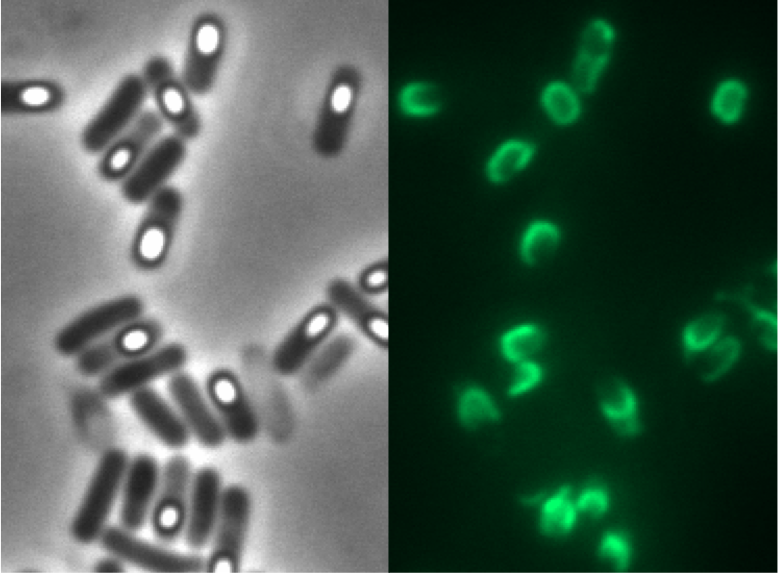
To get a set of promoters with different strength we characterized several promoters in Bacillus subtilis. They can be divided in three different groups: the constitutive promoters from the [http://partsregistry.org/Part:BBa_J23100 Anderson collection] from the Partsregistry, the constitutive promoters PliaG, Pveg and PlepA from B. subtilis, and the inducible promoters PliaI and Pxyl-XylR from B. subtilis. For the characterization of the different promoters we used the lux operon ![]() where gene expression leads to the production of luminescence. For this promoter evaluation the reporter vector pSBBs3C-luxABCDE was used which was not fully in BioBrickStandard in this time because of one last forbidden restriction site. We also used the reporter gene lacZ
where gene expression leads to the production of luminescence. For this promoter evaluation the reporter vector pSBBs3C-luxABCDE was used which was not fully in BioBrickStandard in this time because of one last forbidden restriction site. We also used the reporter gene lacZ ![]() for the promoter evaluation where the promoter activity can be measured by doing β-galactosidase assays. Therefore we used the reporter vector pSBBs1C-lacZ. See this page for an overview and background information of all evaluated promoters and see the Data page for more details.
for the promoter evaluation where the promoter activity can be measured by doing β-galactosidase assays. Therefore we used the reporter vector pSBBs1C-lacZ. See this page for an overview and background information of all evaluated promoters and see the Data page for more details.
Overview of several evaluated promoters
Here you can get an overview of all evaluated promoters which cover a large range of activity. For more details and for informations for the experiments see the Data page of the promoters. Note that Pveg was not evaluated with luminescence measurements and this bar is just projected from the results of the beta-galactosidase assay.
|
 "
"