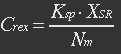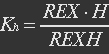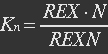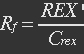Team:Shenzhen/Result/YAO.Sensor
From 2012.igem.org
(Difference between revisions)
| Line 38: | Line 38: | ||
<ul><li>I. Transportation of REX into Mitochondrial Matrix</li></ul> | <ul><li>I. Transportation of REX into Mitochondrial Matrix</li></ul> | ||
<ul><p> We assume that the transcription of SP-REX is constant, SP-REX concentration in cytoplasm is constant, the probably for REX to be transported into each mitochondrion is equal, and mitochondria are of similar size. So with different number of mitochondria in yeast (Nm), the concentration of REX in each mitochondria (Ctf) might differ. Ksp is the transport coefficient of signal peptid which links the XR/Nm with Ctf. For different kinds of signal peptid, Ksp might be different.</p></ul> | <ul><p> We assume that the transcription of SP-REX is constant, SP-REX concentration in cytoplasm is constant, the probably for REX to be transported into each mitochondrion is equal, and mitochondria are of similar size. So with different number of mitochondria in yeast (Nm), the concentration of REX in each mitochondria (Ctf) might differ. Ksp is the transport coefficient of signal peptid which links the XR/Nm with Ctf. For different kinds of signal peptid, Ksp might be different.</p></ul> | ||
| - | [[File:Eqn1.jpg]] (1) | + | <ul><p>[[File:Eqn1.jpg]] (1)</p> |
| - | XSR: SP-REX concentration in cytoplasm. Nm: Number of mitochondria. Crex: Concentration of REX in Mitochondria. Ksp: Transport coefficient of signal peptid. | + | <p><i>XSR: SP-REX concentration in cytoplasm. Nm: Number of mitochondria. Crex: Concentration of REX in Mitochondria. Ksp: Transport coefficient of signal peptid.</i></p></ul> |
| + | |||
| + | <ul><li>II. Activation of REX as a Transcription Factor with NAD+ and NADH</li></ul> | ||
| + | <ul><p> NAD+ and NADH can competitively bind to the same site of REX, when NADH binds to REX, REX will be repressed and cannot bind to ROP. Then the transcription function is repressed. </p></ul> | ||
| + | <ul><p>[[File:Eqn2.jpg]]</p> | ||
| + | <p><i>H stands for NADH, REXH stands for REX bidden with NADH.</i></p></ul> | ||
| + | <ul><p>At equilibrium: [[File:Eqn3.jpg]] (2)</p> | ||
| + | <p><i>N stands for NAD+, REXN stands for REX bidden with NAD+.</i></p></ul> | ||
| + | <ul><p>At equilibrium: [[File:Eqn4.jpg]] (3)</p></ul> | ||
| + | <ul><p>NAD+ and NADH are in a balance, so we assumed the total level of NAD+ and NADH in the mitochondria is constant.</p></ul> | ||
| + | <ul><p>[[File:Eqn5.jpg]]</p></ul> | ||
| + | <ul><p>REX, REXH and REXN are in a balance, so total level of REX, REXH and REXN equals to Crex.</p></ul> | ||
| + | <ul><p>[[File:Eqn6.jpg]] (4)</p></ul> | ||
| + | <ul><p>REX which was not bidden with NADH can bind to ROP.</p></ul> | ||
| + | <ul><p>[[File:Eqn7.jpg]] (5)</p></ul> | ||
| + | <ul><p>REX’ stands for active REX which can bind to ROP.</p></ul> | ||
| + | <ul><p>The fraction of active REX acting as repressor is Rf.</p></ul> | ||
| + | <ul><p>[[File:Eqn8.jpg]] (6)</p></ul> | ||
| + | <ul><p>With (5), (4), (2) and (3) in (6), the fraction becomes</p></ul> | ||
| + | <ul><p>[[File:Eqn9.jpg]] (7)</p></ul> | ||
| + | |||
| + | <div class="figurep"> | ||
| + | [[File:result_yao_sensor_p1.jpg]] | ||
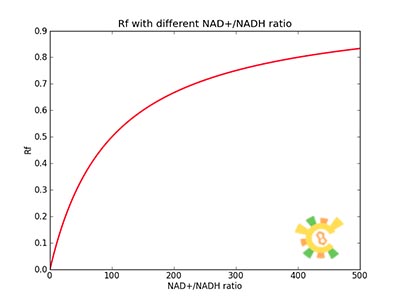
| + | <p>Figure 1. Rf responding to different NAD+/NADH ratio.</p><p>For plotting of this picture the following parameters were used: assuming K_H = 0,02 μM and K_N = 2 μM since REX NADH affinity > REX NAD affinity [2]. </p></div> | ||
| + | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<ul><li>III. Control of promoter activity by REX</li></ul> | <ul><li>III. Control of promoter activity by REX</li></ul> | ||
In yeast mitochondria, transcription initiation factor MTF1, combined with RNA polymerase Rpo41, bind to promoter to initiate transcription. It is MTF1 that discerns the promoter region, and the binding of MTF1 on promoter region is reversible. Also the binding of REX on ROP, a DNA region near promoter is also reversible. | In yeast mitochondria, transcription initiation factor MTF1, combined with RNA polymerase Rpo41, bind to promoter to initiate transcription. It is MTF1 that discerns the promoter region, and the binding of MTF1 on promoter region is reversible. Also the binding of REX on ROP, a DNA region near promoter is also reversible. | ||
Revision as of 12:49, 25 September 2012
 "
"