Team:Bielefeld-Germany/Labjournal/week17
From 2012.igem.org
(Difference between revisions)
(→Monday August 20th) |
(→Wednesday August 22nd) |
||
| Line 90: | Line 90: | ||
**Restarted from the beginning and made a gradient-PCR for [http://partsregistry.org/Part:BBa_K863121 GFP_His] and [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] to get some more product. | **Restarted from the beginning and made a gradient-PCR for [http://partsregistry.org/Part:BBa_K863121 GFP_His] and [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] to get some more product. | ||
* '''Team Cultivation & Purification:''' | * '''Team Cultivation & Purification:''' | ||
| - | ** Made | + | ** Made SDS-Pages of cultivation from 08/15.[[File:Bielefeld2012_0826_3.jpg|2500px|thumb|right|'''Figure 1: Cultivation from [https://2012.igem.org/Team:Bielefeld-Germany/Labjournal/week16#Wednesday_August_15th 08/15]''' of ''E. coli'' KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015], ''E. coli'' KRX (negative control) and ''E. coli'' KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] (positive control). 250 mL in 1 L flasks without baffles, [https://2012.igem.org/Team:Bielefeld-Germany/Protocols/Materials#Autoinduction_medium autoinduction medium] with 60 µg/mL chloramphenicol at 37 °C for 12 hours.]] |
* '''Team Cloning of Bacterial Laccases:''' | * '''Team Cloning of Bacterial Laccases:''' | ||
** Colony PCRs: some prefix insertions of promoters in pSB1C3 with ecol and ecol_HIS showed positive bands, so we picked the colonies for plasmid isolation. The problem with the prefix insertion of the promoters in pSB1C3 with the ecol gene is that we can’t easily prove that the promoters were ligated in the backbone because with control restriction there would be an about 50 bp longer fragment which is difficult to see even in 3 % agarose gel. For this reason we designed FW primers (J23103_K_FW and J23110/117_K_FW and T7_K_FW) for our colony PCR, which can just bind, if the new promoter sequence is included in the plasmid. Until all colonies from the promoter J23110 ligation were red, we had some white colonies for the pT7 promoter in pSB1C3 backbone. So we picked a positive colony and plated it for plasmid isolation. | ** Colony PCRs: some prefix insertions of promoters in pSB1C3 with ecol and ecol_HIS showed positive bands, so we picked the colonies for plasmid isolation. The problem with the prefix insertion of the promoters in pSB1C3 with the ecol gene is that we can’t easily prove that the promoters were ligated in the backbone because with control restriction there would be an about 50 bp longer fragment which is difficult to see even in 3 % agarose gel. For this reason we designed FW primers (J23103_K_FW and J23110/117_K_FW and T7_K_FW) for our colony PCR, which can just bind, if the new promoter sequence is included in the plasmid. Until all colonies from the promoter J23110 ligation were red, we had some white colonies for the pT7 promoter in pSB1C3 backbone. So we picked a positive colony and plated it for plasmid isolation. | ||
Revision as of 15:06, 25 September 2012
Contents |
Week 17 (08/20 - 08/26/12)
Monday August 20th
- Team Cellulose Binding Domain:
- Biggg Colony-PCR of the [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-ligation with the result, that all seem to carry [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]. The ligation didn't work at all...
- Team Cloning of Bacterial Laccases:
- Digestion of pSB1C3 plasmid backbone, tthl_HIS and bpul_HIS for doing the assembly with the new T7 promoter and J23110 again.
- We did PCRs on bpul_HIS, bhal_HIS and ecol_HIS again.
- Team Cultivation & Purification: Today we decided to finish our lean period of non-active laccases. It was time for a transcript analysis to have a look if our laccases are transcribed into RNA and if there is a lot of it or if its gets degraded just in time. We took our cells, centrifuged and shock freezed them in liquid nitrogen to stop every metabolism in the cell and freeze the status of the RNA. The frozen cell pellets were stored at -80°C for the upcoming tests.
- Team Immobilization
- Today we set up a new immobilization experiment following the same procedure as in week 15 (see week 15 Monday August 6th-Wednesday August 8th /Team Immobilization) but with different bead concentrations: 0.06, 0.08, 0.1, 0.12, 0.14 and 0.16 mg/mL. Based on the results of the first experiment, the samples were incubated with TVEL0 for 36h.
Tuesday August 21st
- Team Cellulose Binding Domain:
- Tested restriction enzymes.
- Team Activity Tests: I know, all of you are expecting us to report about some laccase activity now, but today is going to be different. Today we started with a special task: a q-PCR. As written in one of our last labjournal entries we have discussed a lot and searched for reasons why our laccases are not active. We decided that one possible step to find the mistake is to analyze the transcript. The level of mRNA will show us if our plasmid is expressed at all or if there might be something wrong with it. Team Cultivation cultivated small samples of E.coli KRX with plasmids of laccases from the following organisms:Escherichia coli, Bacillus pumilus, Bacillus halodurans C-125, Xanthomonas campestris pv. campestris B100 and Thermus thermophilus. As controls a sample with a [http://partsregistry.org/wiki/index.php/Part:BBa_K525710 ligase a] plasmid (to ensure the induction works) and E. coli KRX (without any plasmid) were used. So our first step today was freezing some cell pellets for a RNA isolation and ordering primers. We will go on with our new mission as soon as the primers arrive.
- Team Cloning of Bacterial Laccases:
- The isolated plasmid from the 15th August was prepared and given up for sequencing.
- We designed primers for q-PCR for Team Activity Tests (details, look above). The PCRs with this primer pairs should result in about 200 bases long fragments.
- We realized that the primer pairs we anneal for the promoter parts had about 2000 ng/µl if diluted 1:10 from originally 100 pmol. Maybe our used amounts on promoter for the assemblies was a LITTLE to high. So now we try a dilution of 1:1000 with about 2 ng/µl.
- PCR products of different laccases were purificated and digested for suffix insertion.
- Ligation of the laccase genes ecol, ecol_HIS and bpul and bpul_HIS, the 1:000 diluted (0,1 pm/µl) promoters (pT7 and P110) in pSB1C3 vector.
- Ligation of the digested ecol_pSB1C3 and ecol_HIS_pSB1C3 plasmids with the 1:000 diluted (0,1 pm/µl) promoters (pT7 and P110).
- Ligation of T7 promoter and J23110 promoter in pSB1C3 backbone.
- Team Fungal and Plant Laccases:
- We designed primers for the laccase <partinfo>BBa_K500002</partinfo> for cloning in P. pastoris shuttle vector.
Wednesday August 22nd
- Street Science:
- Went to Dr. Joe Max Risse today and asked him about the mircoorganisms of the Fermentationgroup. He recommended Euglena gracilis, since it is without risk, big, colorful, and very healthy under the microscop.
- I asked for the bacteria they have and he told me I could check if their Kocuria rosea is a wild type.
- In the DMSZ catalog there are three strains of Kocuria rosea and all have been assessed to be of riskgroup 1, which is the safest group of bacteria and means it is unlikely that it will infect humans.
- I also asked for Penicillium chrysogenum but the one they use is a GVO.
- Team Site Directed Mutagenesis:
- Ordered new primers for Xantomonas Campestris SDM, since there is still no positive colony and even gradient-PCR gave the wrong product
- Team Cellulose Binding Domain:
- Test-restriction of all isolated [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]- and [http://partsregistry.org/Part:BBa_K863121 GFP_His]-plasmids with NotI with no positive result.
- Restarted from the beginning and made a gradient-PCR for [http://partsregistry.org/Part:BBa_K863121 GFP_His] and [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] to get some more product.
- Team Cultivation & Purification:
- Made SDS-Pages of cultivation from 08/15.
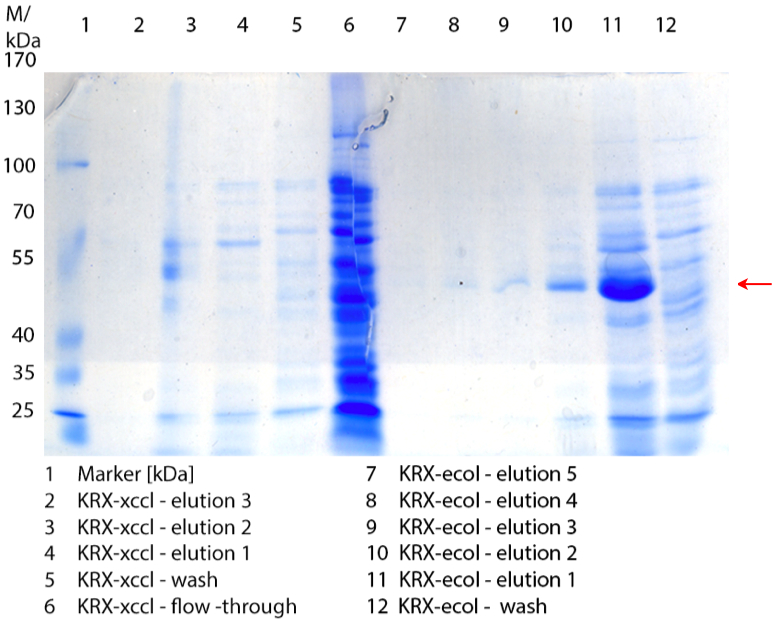 Figure 1: Cultivation from 08/15 of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015], E. coli KRX (negative control) and E. coli KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] (positive control). 250 mL in 1 L flasks without baffles, autoinduction medium with 60 µg/mL chloramphenicol at 37 °C for 12 hours.
Figure 1: Cultivation from 08/15 of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015], E. coli KRX (negative control) and E. coli KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] (positive control). 250 mL in 1 L flasks without baffles, autoinduction medium with 60 µg/mL chloramphenicol at 37 °C for 12 hours.
- Made SDS-Pages of cultivation from 08/15.
- Team Cloning of Bacterial Laccases:
- Colony PCRs: some prefix insertions of promoters in pSB1C3 with ecol and ecol_HIS showed positive bands, so we picked the colonies for plasmid isolation. The problem with the prefix insertion of the promoters in pSB1C3 with the ecol gene is that we can’t easily prove that the promoters were ligated in the backbone because with control restriction there would be an about 50 bp longer fragment which is difficult to see even in 3 % agarose gel. For this reason we designed FW primers (J23103_K_FW and J23110/117_K_FW and T7_K_FW) for our colony PCR, which can just bind, if the new promoter sequence is included in the plasmid. Until all colonies from the promoter J23110 ligation were red, we had some white colonies for the pT7 promoter in pSB1C3 backbone. So we picked a positive colony and plated it for plasmid isolation.
- We plated colonies on new plates, which weren't red from the transformation of pSB1C3 + pT7. We can't do colony PCR on this plasmids, because the resulting parts would be smaller than 100 bps.
Thursday August 23rd
- Team Cloning of Bacterial Laccases:
- Made plasmid isolation of pSB1C3 + pT7 and sent plasmids for sequencing.
- Team Fungal Laccases: We did PCRs on tvel35 plasmid with primer pairs Pc_lac35.P.FW / Pc_lac35.S.RV for trying ligation in pSB1C3 again and with Pc_lac35_FW_oS / Pc_lac35_RV for cloning in shuttle vector.
- Team Cellulose Binding Domain:
- Clean-up of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]- and [http://partsregistry.org/Part:BBa_K863121 GFP_His]-PCR-product (pooled good ones and bad ones, respectively).
- Restriktion of the [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-PCR-product with EcoRI and PstI for ligation with the backbone and EcoRI and AgeI for a assembly.
- Restriktion of the [http://partsregistry.org/Part:BBa_K863121 GFP_His]-PCR-product with EcoRI and PstI for ligation with the backbone and NgoMIV and PstI.
Friday August 24th
- Team Site Directed Mutagenesis:
- Plasmid-isolation and digestion of tvel10 colonies with NotI showed that two of the plasmids have an insert of the right size.
- Team Cellulose Binding Domain:
- Stopped restrictions and clean-up them up. Dephosphorilated the linearized backbone and ligated it to [http://partsregistry.org/Part:BBa_K863121 GFP_His], [http://partsregistry.org/Part:BBa_K863121 GFP_His] and assembled [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His].
- [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]-cells plated to get some more plasmid.
Saturday August 25th
STREET SCIENCE Visit of Danish guys till 08/27
Sunday August 26th
- Team Cellulose Binding Domain:
- Made-up the idea to have a constitutive promoter (like <partinfo>BBa_J61101</partinfo>) in front of the cellulose binding domain and the GFP. With a Freiburg-Prefix in front of the CBDs we could even easily switch directions of CBD and GFP.
- Transformed <partinfo>BBa_J61101</partinfo> from the iGEM-plate into KRX and plated it on AMP-selection-dish.
- Team Cultivation & Purification:
- Made precultures of E. coli KRX without plasmid (negative control) and with plasmids containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015]. As postive control we used [http://partsregistry.org/Part:BBa_K525710 BBa_K525710 (Ligase A)].
| 55px | | | | | | | | | | |
 "
"





