Team:Chalmers-Gothenburg/Results
From 2012.igem.org
m (→Results Summary) |
(→Introduction of indigo synthesizing genes) |
||
| Line 95: | Line 95: | ||
| - | + | ='''Introduction of indigo synthesizing genes'''= | |
| + | <!-- | ||
| + | |||
| + | =='''Construction of pIndigo'''== | ||
| + | To enable production of indigo in yeast, two genes, ''tnaA'' and ''fmo'', had to be cloned into the yeast strain. In order to achieve that, a plasmid containing both genes under the control of two different promoters had to be constructed. First we used PCR to amplify the genes. ''TnaA'', encoding the enzyme tryptophanase, was amplified from ''E. coli'' genomic DNA by colony-PCR and fmo, encoding the flavin-containg monooxygenase, was amplified by PCR from plasmid kindly provided to us by Si Wouk Kim from Chosun University in South Korea. After a few attempts both genes were successfully amplified. TnaA and fmo were digested with ''Bam''HI and ''Xho''I and ''Not''I and ''Pac''I respectively and the genes were separately cloned into pIYC04 which had been digested in the same way as respective gene. The plasmids were now called pIYC04-tnaA and pIYC04-fmo. After the ligation ''E. coli'' DH5α cells were transformed with pIYC04-tnaA and pIYC04-fmo respectively. Both transformations were successful and the plasmids were purified. | ||
| + | |||
| + | Digested ''tnaA'' was ligated with pIYC04-fmo, and digested ''fmo'' was ligated with pIYC04-tnaA and the plasmids were now called pIndigo. The reason that pIndigo was constructed in both ways was in order to increase the chances of getting a functional plasmid. DH5α were transformed with pIndigo. The amplification was successful and pIndigo was purified. pIndigo now contained ''tnaA'' under control of the constitutive ''PGK1'' promoter and ''fmo'' under the control of the constitutive ''TEF1'' promoter. The plasmid was verified by sequencing (data not shown). Sequencing showed that the plasmid contained both ''tnaA'' and ''fmo'' and there were no signs of mutations. | ||
| + | The yeast strain CEN.PK 113-11C was transformed with pIndigo and the presence of the plasmid should theoretically enable the cells to produce indigo, but no blue cells were seen on the plates. To see if the ''tnaA'' and the ''fmo'' gene had been correctly introduced into the cells, a colony-PCR with a following gel electrophoresis was performed for the genes. The expected bands were approximately 1400 bp for both genes. As one can see on the gel picture in Figure 6, bands with right lengths were present for both genes. | ||
| + | |||
| + | Figure 6. Verification of transformation of CEN.PK 113-11C with pIndigo through gel electrophoresis. For both tnaA and fmo was one band expected to be seen with a length of approximately 1400 bp. As one can see on gel picture, bands with right lengths were present for both genes. | ||
| + | |||
| + | After the CEN PK 113-11C had been transformed with the pIndigo the cells were transferred to SD His- plates along with two controls consisting of CEN PK 113-11C without the plasmid and CEN PK 113-11C with empty plasmid. The control cells that lacked the plasmid were not able to grow on the plates, while the transformed ones did, so the transformation was considered to be successful. After more than one week of cultivation in room temperature and 30 ºC no blue cells were detected. After some time the cells were also transferred to fresh SD His- plates, and further cultivated, but this did not give blue cells on the plate. The strains were also cultivated in SD His- media in room temperature, but no blue colour was obtained. | ||
| + | |||
| + | =='''Test with tryptophan added to media'''== | ||
| + | |||
| + | =='''Enzymatic assay of tryptophanase'''== | ||
| + | |||
| + | |||
| + | =='''Test with indole added to media'''== | ||
| + | |||
| + | ==='''Day1'''=== | ||
| + | ==='''Day2'''=== | ||
| + | ==='''Day3'''=== | ||
| + | |||
| + | =='''Absorption spectra'''== | ||
| + | |||
| + | =='''Construction of pFIG1-Indigo'''== | ||
| + | --> | ||
='''Expression of the gene for the LH/CG receptor in yeast'''= | ='''Expression of the gene for the LH/CG receptor in yeast'''= | ||
Revision as of 09:36, 25 September 2012
Contents |
Results Summary
Survival of yeast in urine
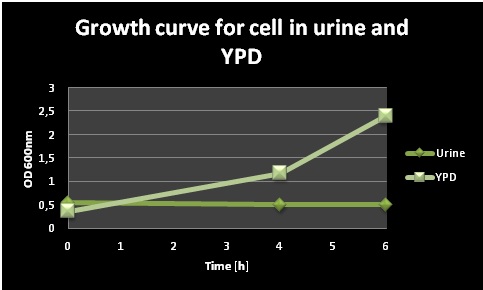
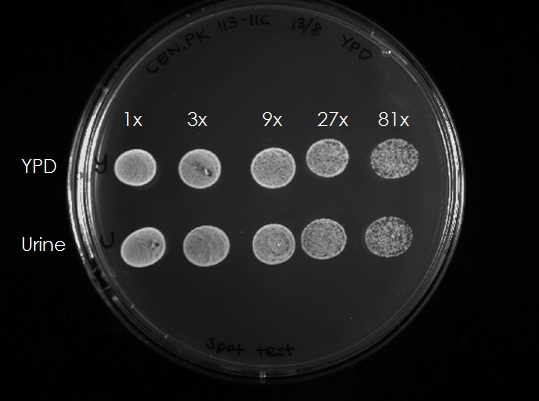
In order to test if the cells remained alive in the urin, 300 µl of each culture (after 4h) were taken and OD600 was adjusted to 0.5. The samples were diluted 3x, 9x, 27x and 81x and 10 µl aliquots of each dilution were spotted on an YPD plate (Figure 2). The cells from both the YPD and the urine media could grow, this means that the cells survived 4 h in urine and grew normal after being spotted on a YPD plate. Summarizing, yeast cells are unable to proliferate in the urine medium but do survive under these conditions.
Deletion of CWP2 gene
Gene Deletion
One task of our iGEM project was the deletion of the CWP2 gene, which is encoding a cell wall mannoprotein. By removing it, we aimed for higher cell wall permeability and thus enhanced chances of our ligand hCG to pass the cell wall and to bind to the membrane-bound receptor. The gene deletion was performed according to the bipartite method. The results from the first PCR reactions, in which we amplified the overlapping fragments, can be seen in Figure 3.
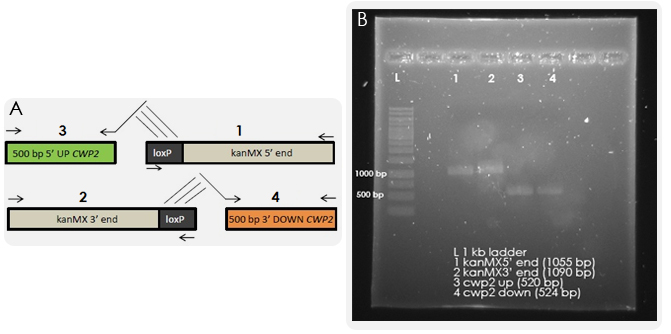
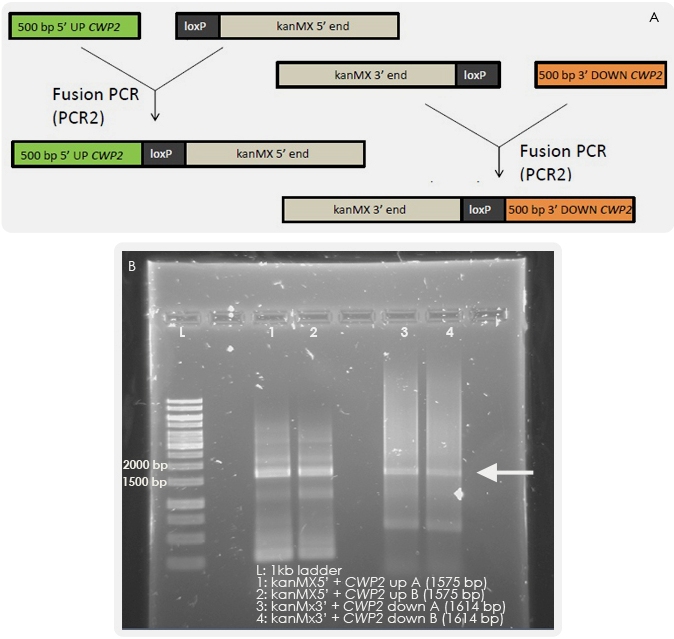
The next step was to fuse together the overlapping fragments. The results of the fusion PCR are shown in the Figure 4. By transforming yeast with these fragments, homologous recombination should occur which will lead to the exchange of the CWP2 gene with the kanMX cassette.
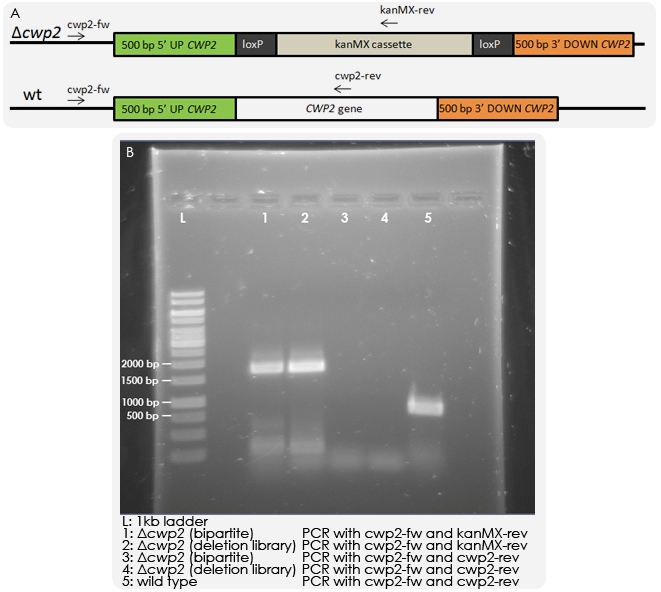
Colony PCR
Yeast strain IMFD-73 was transformed with 150 ng of each fragment and grown on G418 plates. Colonies could be observed, which were purified, and colony PCR was then performed in order to test whether the kanMX cassette was integrated randomly in the genome or had replaced the CWP2 gene as desired. PCR was run with a forward primer outside of the inserted region and reverse primers within the CWP2 gene or kanMX cassette respectively. As a control, a wild-type strain and a Δcwp2 strain from a deletion library were also tested. The results of the PCR reactions indicated that the deletion was correct (see Figure 5).
Lyticase assay
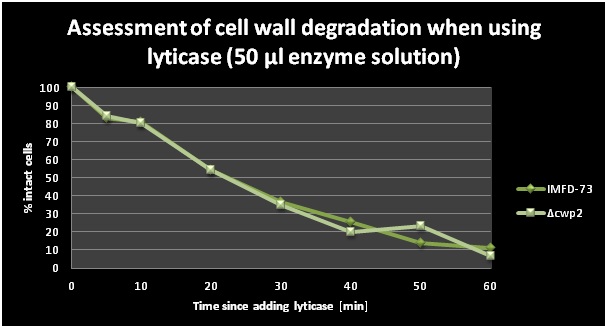
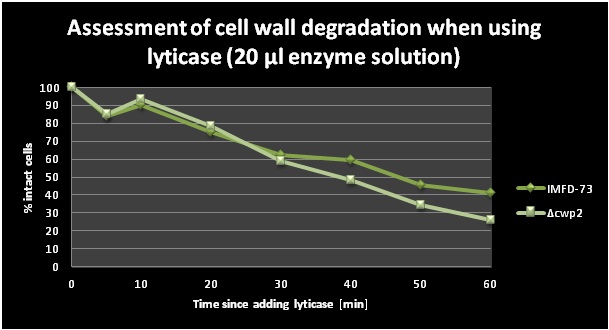
A lyticase assay was performed primarily in order to check the activity of lyticase. Another goal was to compare the rapidity of the cell wall degradation between the IMFD-73 and the [IMFD-73 Δcwp2::kanMX] strain. Lyticase was added to the cells and the cell wall degradation (i.e. protoplast formation) could be displayed by adding SDS and then measuring OD at 600 nm (SDS leads to lysis of protoplasts, while cells with intact cell wall remain unaffected). SDS was added and OD values were taken at different time points. The two graphs (Figure 6 and 7) below show the decrease of the amount of intact cells in % over a time period of 1h with two different amounts of lyticase. The percentages of intact cells were calculated as OD600(t=x)/OD600(t=0)*100. One can observe that a slightly smaller amount of cells of the deletion strain were intact between 30 and 60 minutes (except for one outlier at 50 min) which means that the cell wall degraded quicker in the [IMFD-73 Δcwp2::kanMX] strain. This lead to the conclusion that the cell wall is somewhat weakened in the strains with the deleted cell wall mannoprotein gene CWP2.
Introduction of indigo synthesizing genes
Expression of the gene for the LH/CG receptor in yeast
Two different versions of the LH/CG receptor were to be evaluated and expressed in yeast during this project: one with the original human signal peptide fused to the receptor and one with the yeast Ste3 signal peptide fused to the receptor. Both versions were obtained using the gene construct illustrated in Figure 24. The construct was ordered from and synthesized by GenScript.
Obtaining and cloning the yLHCGR and the hLHCGR gene
The yeast signal peptide version of the receptor gene, hereafter referred to as “yLHCGR”, was obtained with PCR using the original gene construct (Figure 24) and 5’ phosphorylated primers (lhcgr-rev and lhcgr-fw), as can be seen in Figure 25 A. The yLHCGR construct was then religated and when cut with restriction enzymes NotI and PacI and run on a gel, the obtained band displayed a correct length of approximately 2100 bp (see Figure 25 C). The band was cut out and the yLHCGR gene was then ligated into the shuttle plasmid pSP-GM1 and amplified in E. coli.
The human signal peptide version of the receptor gene, hereafter referred to as “hLHCGR”, was obtained by digesting the original gene construct with NotI and PacI (Figure 25 B). Gel electrophoresis was run and the obtained fragment displayed a correct length of approximately 2100 bp (see Figure 25 C). The band was cut out, ligated into the pSP-GM1 plasmid and amplified in E.coli.
Saccharomyces cerevisiae strains IMFD-70, IMFD-72, IMFD-73 and IMFD-74 were then successfully transformed with the yLHCGR and hLHCGR expression plasmids respectively. All these strains have been optimized for the coupling of human GPCRs to the pheromone signaling pathway and they differ in their expression of different G alpha proteins. IMFD-70 has the endogenous G alpha protein [KÄLLA] while the other strains express different versions of yeast/human chimeric G alpha proteins. These chimeric proteins have had the five last amino acid residues of the C terminal end replaced with the corresponding human amino acids [Unpublished data]. IMFD-73 has a chimeric alpha subunit of such type that it should have the best chances of interacting with the LH/CG receptor and was therefore the strain that was mainly used during this project. All strains were kindly provided by the Kondo group at Kobe University, Japan.
Sequencing
In order to confirm that the yLHCGR gene and the hLHCGR gene not only have the correct length but also the correct sequence, the yLHCGR and the hLHCGR plasmid were sent in for sequencing. Results indicated that the sequences were correct for both genes.
Evaluating the functionality of the receptor
The ability of the receptor to detect hCG (i.e to bind the hormone and transmit the signal) when expressed in yeast, was evaluated several times under different conditions. Initial tests were performed by screening for fluorescence after the addition of hCG to the yeast cells. A final test also included screening for indigo production in the completed biosensor system.
Initial tests: Screening for GFP expression
The yeast strains that have been transformed with the gene for the LH/CG receptor have the gene for GFP placed under the control of the pheromone-induced FIG1 promoter. An initial test to see if the LH/CG receptor could detect hCG and activate the pheromone pathway, was therefore done by screening for fluorescence. Below, results from three of these screens are presented.
IMFD-70, IMFD-72, IMFD-73 and IMFD-74, grown at 30°C
One experiment was carried out using all four, IMFD-70, -72, -73 and -74 strains, transformed with yLHCGR, hLHCGR and the empty pSP-GM1 plasmid respectively. Cells were cultivated at 30°C and were treated with lyticase in order to degrade the cell wall. 200 IU of hCG was then added to each strain. Screening for fluorescence was done using a fluorescence microscope. See Figure 3, 4, 5 and 6 for pictures that have been selected as representative of all results obtained during this experiment.
Results obtained for the cells not treated with hCG and cells transformed with the negative control plasmid, indicate that there is relatively high level of background noise of GFP expressions in all IMFD strains. Generally, the fluorescence intensity is approximately equal between samples that have had the hCG hormone added to them and samples that did not have hCG added to them. It was therefore concluded that the LH/CG receptor, when expressed in these yeast strains and tested under the given conditions, is unable to detect hCG or at least that the receptor it is not functioning in a way that produces a sufficiently strong GFP output signal to be distinguished from the background noise.
IMFD-73, grown at room temperature
In a second test, the transformed IMFD-73 strain with either the yLHCGR, hLHCGR or the empty pSP-GM1 plasmid, were cultivated at room temperature. Gron wth at lower temperatures is often used in heterologous protein production since it may improve protein folding. Cells were then treated with cell wall-degrading enzyme lyticase and 200 IU of hCG was added. Results from the fluorescence microscope can be seen in Figure 30, 31 and 32.
Once again, high levels of background fluorescence were present. Results indicated that also when cells are grown at room temperature, the LH/CG receptor is unable to detect hCG or uncapable to produce an output signal that can be distinguished from the background noise in response to detecting hCG.
IMFD-73-ΔCWP2, grown at 30°C
In a third test, the deletion strain IMFD-73-ΔCWP2 transformed with yLHCGR, hLHCGR and the empty pSP-GM1 plasmid respectively was used. The IMFD-73-ΔCWP strain potentially has a weakened cell wall. Cells were grown at 30°C and were not treated with lyticase before the addition of 200 IU hCG. Fluorescence microscope pictures can be seen in Figure 33, 34 and 35.
As previously, the yeast transformed with the LH/CG receptor gene and that had the hCG hormone added to them, did not produce an output signal that could be distinguished from the fluorescence intensity of cells that did not have hCG added to them or that did not contain the LH/CG gene.
Final test of the completed system: screening for color change
One last test was performed using the completed biosensor system: IMFD-73-ΔCWP2+pFig-Indigo+hLHCGR or yLHCGR. The cultures were grown over night before adding hCG in the morning. 3 hours later more hCG was added. From the colorometic assay (see "Results: indigo"), it was concluded that it takes approximately 7 hours for the pIndigo cells to produce a visible color change when grown at 30°C. However, 7 hours after adding hCG to the finalized system, no color change could be observed in any of the cultures (see Figure 36 and 37) and it was concluded that the biosensor did not succeed in detecting the hormone.
 "
"