Team:HokkaidoU Japan/Notebook/plastic Week 9
From 2012.igem.org
| Line 104: | Line 104: | ||
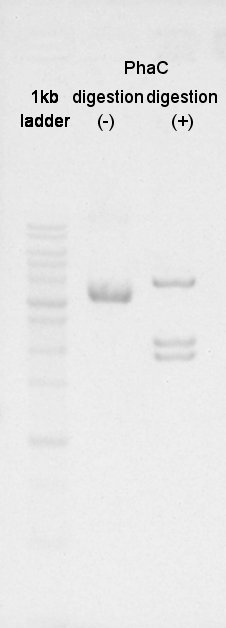
[[image:HokkaidoU2012 120827 phaC digestion sugama.jpg|thumb|digestion result]] | [[image:HokkaidoU2012 120827 phaC digestion sugama.jpg|thumb|digestion result]] | ||
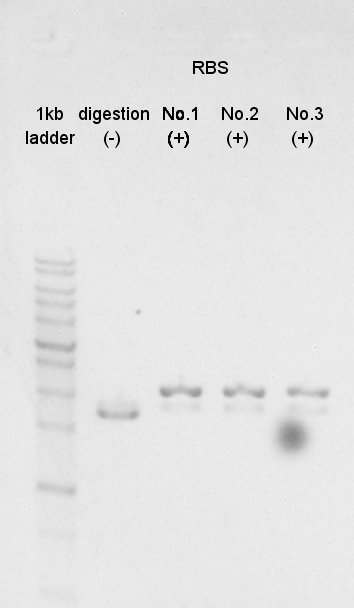
| + | [[image:HokaidoU2012 120827 RBS digestion sugama.jpg|thumb|digestion result]] | ||
| + | |||
</p> | </p> | ||
Revision as of 06:24, 24 September 2012
August 27th
Digestion
Digestion to divide BBa_K342001(PhaC) with XbaI and PstI.
And BBa_B0034(RBS) with SpeI and PstI (with three samples).
PhaC (BBa_K342001)
| DNA solution (100 ng/ul) | 12.5 ul |
| XbaI | 1 ul |
| PstI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 3.5 ul |
| Total | 20 ul |
RBS (BBa_B0034)
N0.1
| DNA solution (20.3 ng/ul) | 14.3 ul |
| SpeI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2.5 ul |
| DW | 0.2 ul |
| Total | 25 ul |
N0.2
| DNA solution (15.6 ng/ul) | 18.6 ul |
| SpeI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2.5 ul |
| DW | 1.9 ul |
| Total | 25 ul |
N0.3
| DNA solution (16.9 ng/ul) | 17.2 ul |
| SpeI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2.5 ul |
| DW | 3.3 ul |
| Total | 25 ul |
Electrophoresis
We confirmed success of digestion by electrophoresis.
The DNA was extracted from TBE gel and we got DNA solution.
Gel extraction
Gel extraction for digestion product. Used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
August 28th
Digestion
Digestion to divide PhaA and PhaB with XbaI and PstI.
I digested PhaC(BBa_K342001) with these restriction sites and also XhoI to divide pSB1C3 into pieces, on which PhaC is.
Because the length of pSB1C3 is nearly PhaC.
And I digested PhaC(BBa_K342001) and pSB1C3 with XbaI and SpeI.
PhaA
| DNA solution (125 ng/ul) | 6.6 ul |
| XbaI | 1 ul |
| PstI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 9.4 ul |
| Total | 20 ul |
PhaB
| DNA solution (125 ng/ul) | 4 ul |
| XbaI | 1 ul |
| PstI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 12 ul |
| Total | 20 ul |
PhaC(BBa_K342001)
| DNA solution (125 ng/ul) | 10 ul |
| XbaI | 1 ul |
| PstI | 1 ul |
| XhoI | 5.1 ul |
| 10xM buffer | 2 ul |
| DW | 0.9 ul |
| Total | 20 ul |
Electrophoresis
We confirmed success of digestion by electrophoresis.
The DNA was extracted from TBE gel and we got DNA solution.
Gel extraction
Gel extraction for digestion product. Used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
August 29th
PHB polymer ethanolysis
We did ethanolysis of PHB polymer for 4hrs with sample 2~8
Preparation for GC/MS
We did the preparation for GC/MS with sample 2~8.
PCR
We multiplied pSB1C3 by PCR.
Used two different DNA polymerase, KOD-Plus-Neo and KAPA Taq.
| Solution | Volume(ul) |
| DNA | 1 |
| Suffix-EX | 1 |
| Prefix-PS | 1 |
| MgSO4 | 3 |
| dNTP | 5 |
| 10x KOD-Plus-Neo Buffer | 5 |
| KOD-Plus-Neo | 1 |
| DW | 33 |
| Total | 50 |
| Number | Degree | Second |
| 1 | 94 | 120 |
| 2 | 98 | 10 |
| 3 | 63.7 | 30 |
| 4 | 68 | 60 |
| 5 | 4 | HOLD ↑STEP DOWNに書き換える |
Cycle:2~4 x 35
| Solution | Volume(ul) |
| DNA | 1 |
| Suffix-EX (10uM) | 2 |
| Prefix-PS (10uM) | 2 |
| KAPA Taq | 25 |
| DW | 20 |
| Total | 50 |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 63.7 | 30 |
| 4 | 72 | 180 |
| 5 | 4 | HOLD |
Cycle:2~4 x 35
August 30th
EtOH precipitation
Diegested phaA(X/S) and RBS(S)was performed EtOH precipitation.
Ligation
PhaA and RBS, PhaB and RBS, PhaB and pSB1C3 were ligated each other.
And the DNA were transformed into bacteria.
Colony PCR
The length of PhaB on pSB1C3 was confirmed by colony PCR.
The result showed PhaB and pSB1C3 didn't ligate correctly.
Liquid Culture
Cultivation of bacteria holds RBS (BBa_B0034) - PhaC (K342001) was started.
August 31th
Colony PCR
We confirmed the length of the three constructs that transformed yesterday.
The result showed RBS and PhaA were ligated correctly.
So the cultivation was started.
plasmid extraction
Plasmid of RBS-PhaC were extracted.
And then we got 50ul DNA solution.
Liquid culture
Cultivation of bacteria holds dT (BBa_B0015) was started.
September 1st
plasmid extraction
Plasmid of RBS-PhaA and dT (BBa_B0015) were extracted.
And then we got 50ul DNA solution.
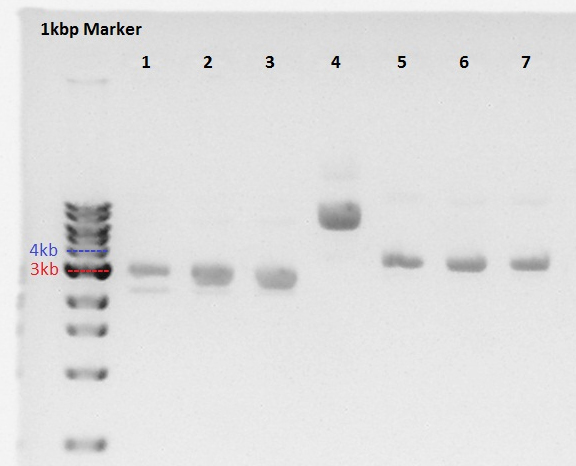
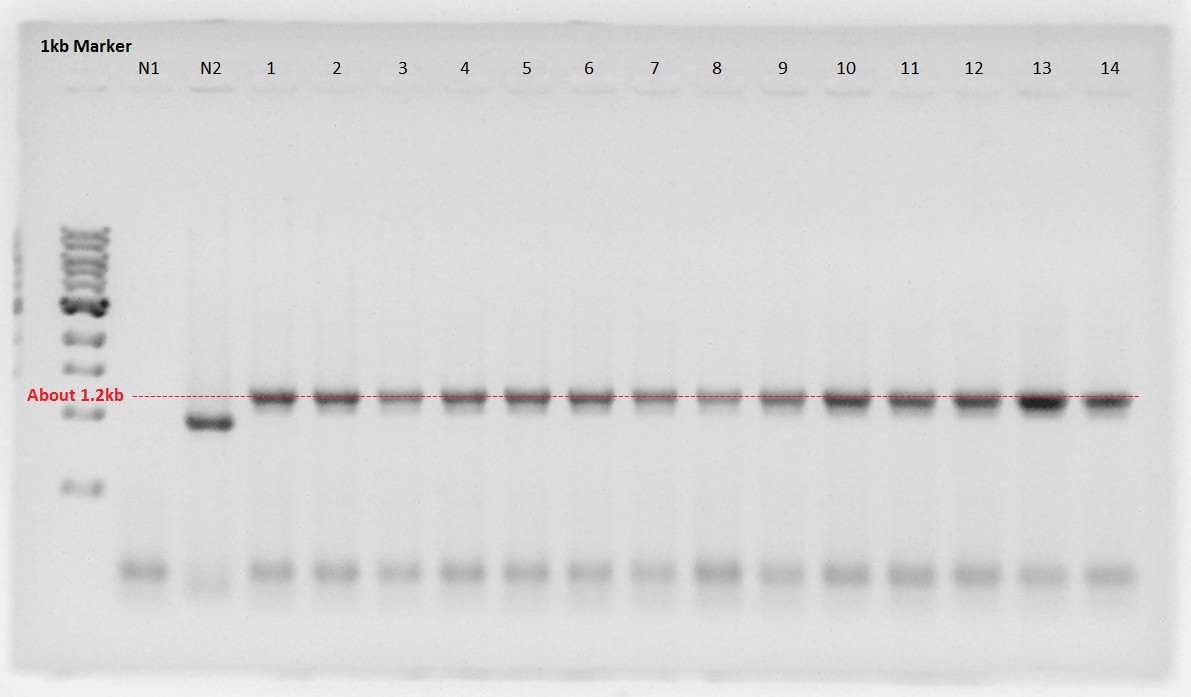
1: dT on pSB1AK3 (About 3.3kbp)
2 to 4: RBS-PhaA on pSB1A2 (About 3.2kbp)
5 to 7: RBS-PhaC on pSB1A2 (About 4.1kbp)
We thought sample4 is not ideal plasmid and trashed it.
Digestion
RBS-PhaA was digested with XbaI and SpeI restriction site to ligate with RBS-PhaC digested with SpeI site.

1 and 2 is digested RBS-PhaA on pSB1A2.
Upper fragment is vector, pSB1A2.
Lower one is an objective fragment, RBS-PhaA (About 1.2kbp).
And PhaB and pSB1C3 were digested with XbaI and SpeI site.
We decided to try ligation PhaB with pSB1C3 again.
Electrophoresis
We confirmed success of digestion by electrophoresis.
The DNA was extracted from TBE gel and we got DNA solution.
September 2nd
EtOH precipitation
The digested DNAs, RBS-PhaA (No.1), RBS-PhaA (No.2), RBS-PhaC on pSB1A2, PhaB and pSB1C3 were concentrated by EtOH precipitation.
Ligation
RBS-PhaA (No.1 and No.2) was ligated with RBS-PhaC on pSB1A2.
And PhaB was taken in pSB1C3.
Transformation
These ligated DNAs transformed into E.coli (strain: DH5α).
And then we spread fungus liquid added LB on LB plates include antibiotics.
 "
"