Team:Tokyo Tech
From 2012.igem.org
(→Ⅱ-2 Organic synthesis of PHA) |
(→Ⅱ-2 Organic synthesis of PHA) |
||
| Line 104: | Line 104: | ||
===Construction of pha-C1-A-B1 in Biobrick format=== | ===Construction of pha-C1-A-B1 in Biobrick format=== | ||
[[File:tokyotech PHA biobrick.png|200px|thumb|right|fig2,]] | [[File:tokyotech PHA biobrick.png|200px|thumb|right|fig2,]] | ||
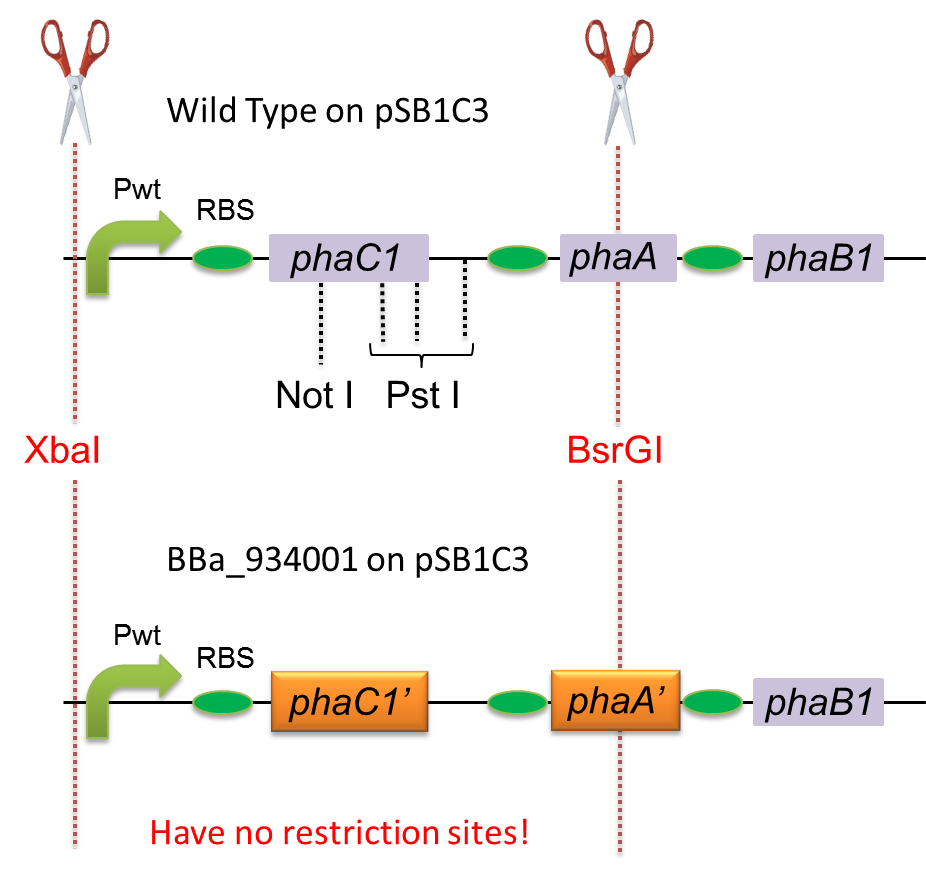
| - | To construct a part that meets Biobrick format, we have modified the pha-C1-A-B1 operon not to contain forbidden restriction enzyme sites. First, we cloned the wild type gene pha-C1-A-B1 from R.eutropha H16 by using PCR and inserted the gene into pSB1C3. However, wild type pha-C1-A-B1 gene sequence contains one NotI and three PstI recognition sites that are not allowed in Biobrick format. To get pha-C1-A-B1 sequence without these recognition sites, we ordered the chemically synthesized DNA from IDT/MBL. In this chemically synthesized DNA, coding is optimized for E.coli. That is to say, we got PHB synthesizing gene in Biobrick format([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]). | + | To construct a part that meets Biobrick format, we have modified the pha-C1-A-B1 operon not to contain forbidden restriction enzyme sites. First, we cloned the wild type gene pha-C1-A-B1 from R.eutropha H16 by using PCR and inserted the gene into pSB1C3. However, wild type pha-C1-A-B1 gene sequence contains one NotI and three PstI recognition sites that are not allowed in Biobrick format. To get pha-C1-A-B1 sequence without these recognition sites, we ordered the chemically synthesized DNA from IDT/MBL. In this chemically synthesized DNA, coding is optimized for <I>E.coli</I>. That is to say, we got PHB synthesizing gene in Biobrick format([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]). |
<br><br><br><br> | <br><br><br><br> | ||
Revision as of 16:01, 22 September 2012
Project over view
A love story contains several processes. Two people fall in love and their love burning wildly. However, no forever exists in the world, in most occasions, love will eventually burn to only a pile of ashes of the last remaining wind drift away. In our project, we have recreated the story of "Romeo & Juliet" by Shakespeare vividly by two kinds of Escherichia coli. We aim to generate a circuit involving regulatory mechanism ofⅠ cell-cell communication
"Romeo and Juliette" is the dramas by dramatist William Shakespeare of England. The stage is city Verona, Italy in the 14th century. Romeo met Juliette. Two persons fall to love instantly. In this project, we will recreate the love story of "Romeo & Juliette".
Ⅰ-1 story
We make our cute E.coli play “Romeo and Juliet” which is one of Shakespeare’s most famous plays. In this project, we define the signal that E.coli produce as the romantic feeling of Romeo and Juliet. In this project, we will recreate the love story of "Romeo&Juliette", by using "Cell-cell communication"
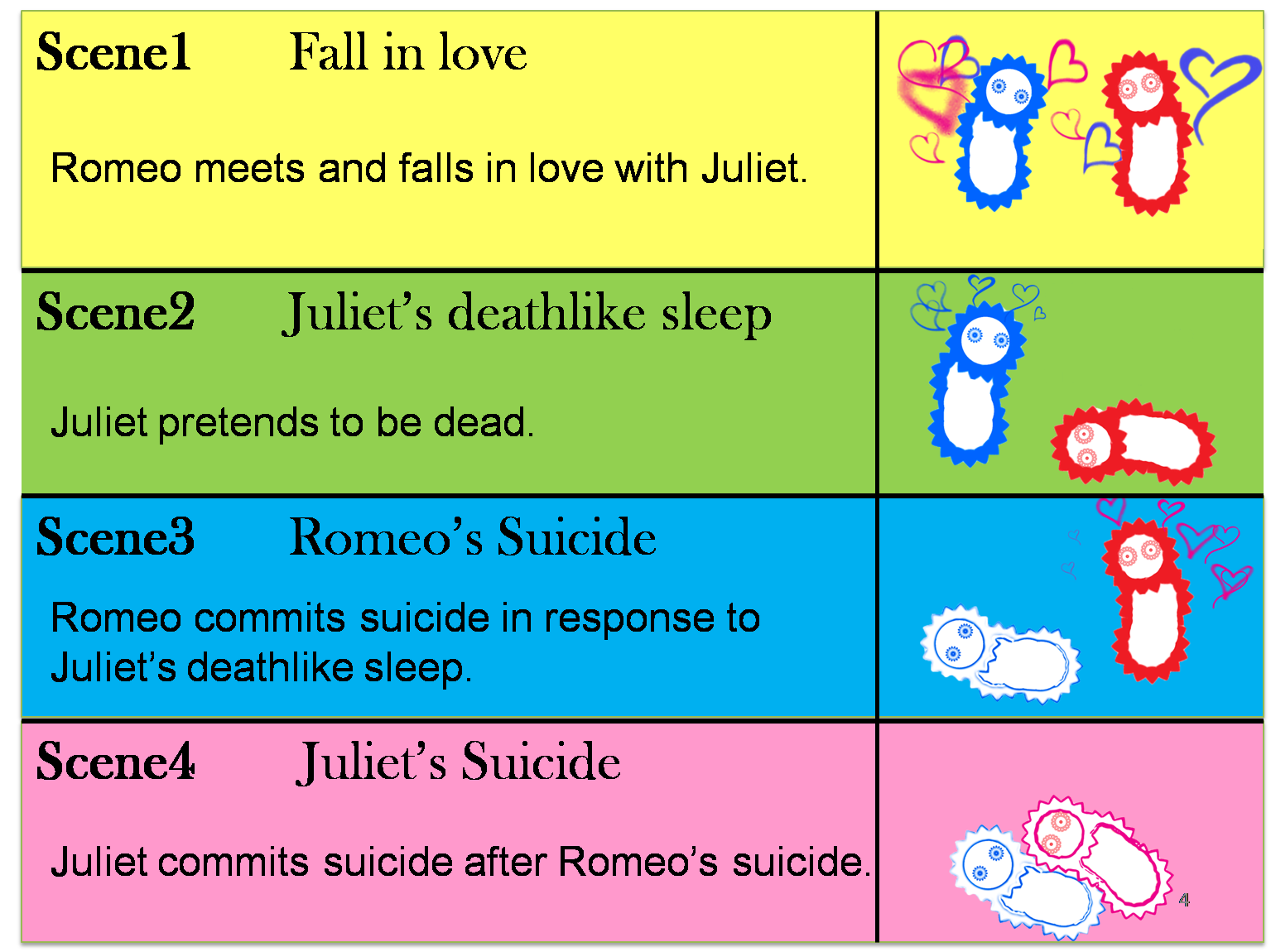
The story that we reproduce is divided into four scenes.
(Scene 1) Romeo and Juliet encountered and they fell in love with each other.
(Scene 2) However, providence lane person, this period of pure love might never broom since they were born in two feuding families. In order to keep their relationship, Juliet planned to pretend die by drinking the drug that will put her into a death like coma so that when she woke up there will be no obstacle between their great loves.
(Scene 3) However, this new was not conveyed to Romeo in time. Romeo misunderstood Juliet’s death and committed suicide.
(Scene 4) When Juliet woke up, what waiting for her was not a beautiful future but Romeo’s death that is waiting for her. Juliet felt so sad and she followed Romeo by stabbing herself with a dirk.
Desingn of our circuit for project.
| Gene Circuit |
We designed a cell-cell communication system that makes two types of engineered E.coli play “Romeo and Juliet”. We represented the story with concentration of signal molecules 3OC6HSL and 3OC12HSL. 3OC6HSL is synthesized by LuxI enzyme in Romeo cell, and 3OC12HSL is synthesized by LasI enzyme in Juliet cell. First, to represent their burning love, we designed a positive feedback system in which the production of a signal activates the production of the other signal. Second, we applied a 3OC6HSL-dependent band detect system to represent the deathlike coma of Juliet cell by a stop of 3OC12HSL production. When the concentration of 3OC6HSL reaches higher level by the positive feedback, the concentration of TetR is enough level to repress the expression of LasI, and the production of 3OC12HSL is stopped. Third, to realize “Romeo suicide”, we designed 3OC12HSL dependent LacI inverter. In the presence of 3OC12HSL, LacI represses the expression of the suicide gene. When Juliet cell is in deathlike coma, supply of 3OC12HSL is stopped, so the suicide gene is expressed and Romeo cell die. Finally, to realize “Juliet suicide”, we also designed 3OC6HSL dependent LacI inverter. After the death of Romeo cell, supply of 3OC6HSL is stopped, and the expression of LacI which represses the suicide gene is stopped. As a result, suicide gene is expressed and Juliet cell die.
Ⅰ-2 Positive Feedback of cell-cell communication
Other experiments for basis of positive feedback system
Detailed characterization of Lux-Tet Hybrid promoter: Key part for band detect
Ⅰ-3 Modeling
Text
Ⅱ Organic synthesis of PHA
Ⅱ-1 Story
There is what of the famous scene of "Romeo and Juliette"
JULIET: O Romeo, Romeo! why are you Romeo? Deny your father and refuse your name; Or, if you will not, be but sworn my love, And I'll no longer be a Capulet.
ROMEO: Shall I hear more, or shall I speak at this?
JULIET: It's but your name that is my enemy; you are yourself, though not a Montague. What's Montague? it is nor hand, nor foot, Nor arm, nor face, nor any other part Belonging to a man. O, be some other name! What's in a name? that which we call a rose By any other name would smell as sweet.
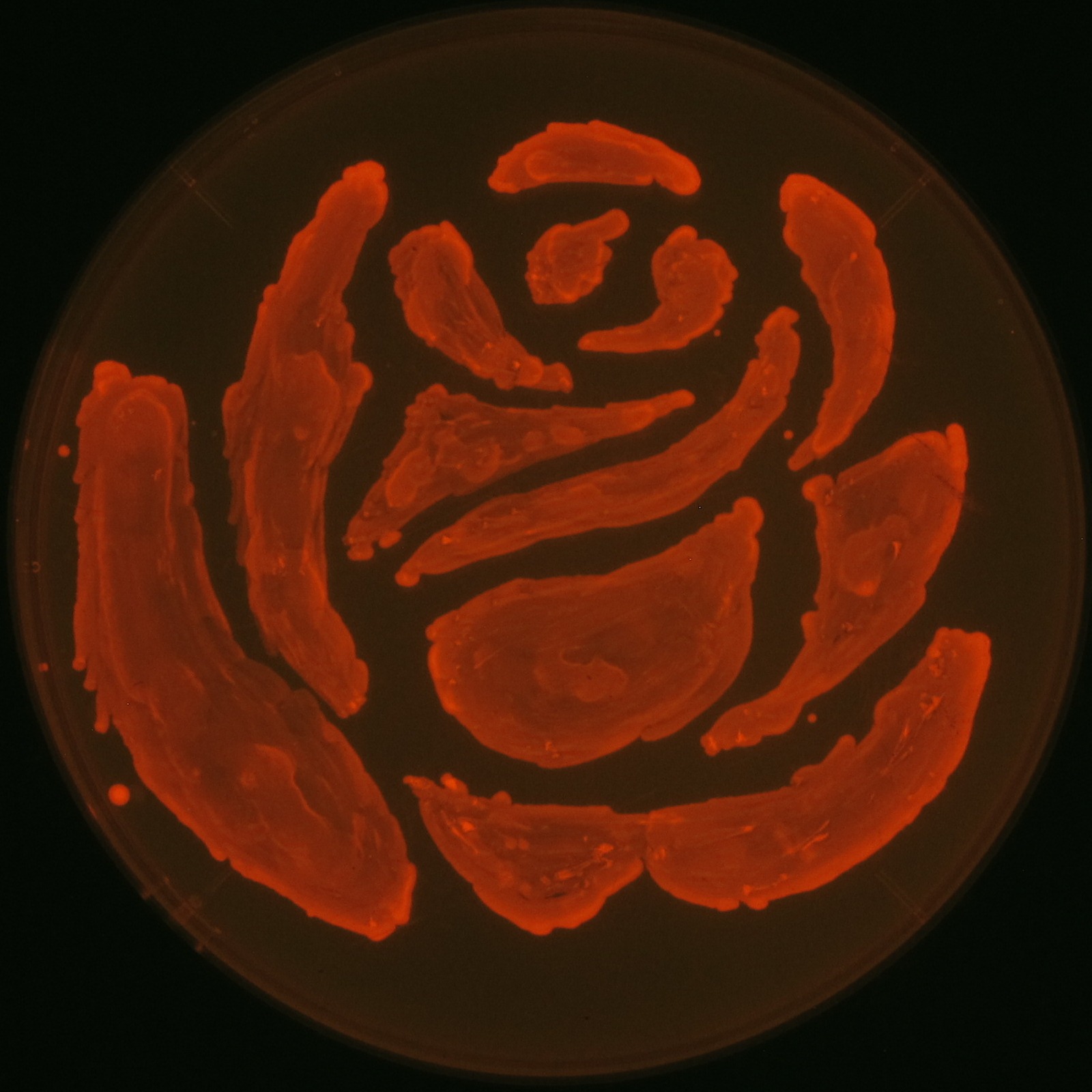
we will recreate the rose come out in the lines of the famous drama'Romeo and Juliet' by the synthesis of PHA.
Ⅱ-2 Organic synthesis of PHA
We made a new biobrick part and succeeded synthesizing Polyhydroixyalkanoates(PHAs). In our project, we designed rose silhouette to enhance the balcony scene of “Romeo and Juliet” by the synthesis of PHAs.
Construction of pha-C1-A-B1 in Biobrick format
To construct a part that meets Biobrick format, we have modified the pha-C1-A-B1 operon not to contain forbidden restriction enzyme sites. First, we cloned the wild type gene pha-C1-A-B1 from R.eutropha H16 by using PCR and inserted the gene into pSB1C3. However, wild type pha-C1-A-B1 gene sequence contains one NotI and three PstI recognition sites that are not allowed in Biobrick format. To get pha-C1-A-B1 sequence without these recognition sites, we ordered the chemically synthesized DNA from IDT/MBL. In this chemically synthesized DNA, coding is optimized for E.coli. That is to say, we got PHB synthesizing gene in Biobrick format([http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]).
Significance in SynBio
cell-cell communication
Ecosystem
Organic synthesis of PHA
Water-repellent
 "
"