Team:British Columbia/Pathway
From 2012.igem.org
(Difference between revisions)
(Created page with "{{Template:Team:British_Columbia_Header}}") |
|||
| Line 1: | Line 1: | ||
{{Template:Team:British_Columbia_Header}} | {{Template:Team:British_Columbia_Header}} | ||
| + | <html> | ||
| + | <style> | ||
| + | #break {width:950px;float:left; background-color: white; margin-left: 8px; margin-top:10px;}</style> | ||
| + | <div id=break></div> | ||
| + | |||
| + | <p align=center><font face=arial narrow size=4><b>Pathway Model | ||
| + | </b></font></p><font face=arial narrow> | ||
| + | |||
| + | The study of environmental genomics attempts to capture the taxonomic and functional diversity of natural microbial communities. Our host at UBC, the Hallam lab, designs novel tools for analyzing the gene content in the context of distributed metabolism. Recently, a pipeline has been developed for the automated construction and visualizing of metabolic pathways from genomic data by integrating software such as Pathway Tools, Pathologic and Metacyc [1]. This provided an opportunity to model pathway compartmentalizing and distribution between microbes in the natural environment as it applies to our project. </br></br> | ||
| + | |||
| + | <b>Summary of the pipeline for community-level metabolic analysis (Figure 1):</b> First, open reading frames predicted from sequence data using Prodigal are annotated by protein BLAST and summarized in a GenBank file. Pathway/genome databases (PGDBs) are generated from sequence data in a manner that does not constrain predictions within the scope of model organisms. This allows for a community-based analysis that can be visualized using Pathway Tools. | ||
| + | |||
| + | <p align=center><img src="https://static.igem.org/mediawiki/2012/9/9c/UbcigemSlide1.jpg"></p> | ||
| + | |||
| + | Modeling Metabolism: Rhodococcus erythropolis and Pseudomnas fluorescens are often both prevalent in similar niches. With a high likelihood that these organisms encounter each other, studies have been conducted to assess their metabolic properties in co-culture. In a study by Kayser et al, it was shown that the 4-S biodesulferization pathway in R.erythropolis was more active in the presence of P.fluorescens [2]. As P.fluorescens does not biodesulferize DBT and sulfate is known to repress the 4-S pathway, we looked to analyze the genomes of the two organisms in the context of sulfur metabolism. </br></br> | ||
| + | |||
| + | The 4-S pathway releases sulfite, which is toxic to the cell, therefore genomes were analyzed with the aforementioned pipeline for pathways involved in metabolizing sulfite. It was found that both organisms have annotated genes converting sulfite into sulfate via a reductase; however, the organisms differ on the downstream meatabolism of the sulfate. Where both organisms have an assimilatory sulfate metabolism, only P.fluorescens has a dissimilatory sulfate metabolism (Figure 3, 4). Based on these findings, we can hypothesize that R.erythropolis and P.fluorescens likely excrete and catabolize any excess sulfate, respectively. This provides an explanation for the improved desulfurization found in co-culture conditions. P.fluorescens potentially removes sulfate from the environment allowing increased secretion by R.erythropolis and thereby removing suppression of the 4-S pathway. The prediction of distributed sulfite metabolism to improve biodesulferization in the environment provides both a testable hypothesis and direction in improving biodesulferization through synthetic pathway distribution. </br></br> | ||
| + | |||
| + | |||
| + | <p align=center><img src="https://static.igem.org/mediawiki/2012/4/43/UbcigemerSlide2.jpg"></p> | ||
| + | <p align=center><img src="https://static.igem.org/mediawiki/2012/d/d1/UbcigemerSlide3.jpg"></p> | ||
| + | |||
| + | |||
| + | We then looked through the literature for more evidence of shared metabolism between both R.erythropolis and P.fluorescens to further analyze gene content in the context of co-culture experiments. In a study by Goswami et al. the metabolism of chlorinated aromatic compounds and phenol was compared in monoculture and co-culture with both R.erythropolis and P.fluorescens [3]. This study showed the growth rate of pure culture R.erythropolis was higher than P.fluorescens on chlorinated aromatics; however, in mixed culture, P.fluorescens showed a higher growth rate. For the degradation of phenol, R.erythropolis showed higher growth rates in both pure and mixed culture. The authors of this study suggested that these results were likely a product of substrate competition. We attempted to analyze the genomes of both R.erythropolis and P.fluorescens, separately and together in an attempt to offer an alternate interpretation of the co-culture results. The first pathways assessed were those involved in chlorinated aromatic degradation. It was found that P.fluorescens contains a higher diversity of genes involved in catabolizing chlorinated aromatics; however, only R.erythropolis seems to be able to degrade phenol (Figure 5 and 6). This suggests the possibility of the compartmentalization of different components of these metabolic processes lead to different growth kinetics seen in co-culture. For example, while R.erythropolis may be more efficient at degrading certain chlorinated aromatics, in co-culture, the diversity of catabolism of chlorinated aromatics allows P.fluorescens to grow more rapidly. However, chlorinated aromatic degradation by P.fluorescens would result in the accumulation of some downstream products, such as phenol, that only R.erythropolis can catabolism. This provides a metabolic network which could both select and sustain both microbes in the presence of diverse chlorinated aromatics. </br> | ||
| + | |||
| + | |||
| + | <p align=center><img src="https://static.igem.org/mediawiki/2012/b/bd/UbcigemSlide4.jpg"></p> | ||
| + | <p align=center><img src="https://static.igem.org/mediawiki/2012/9/99/UbcigemSlide5.jpg"></p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
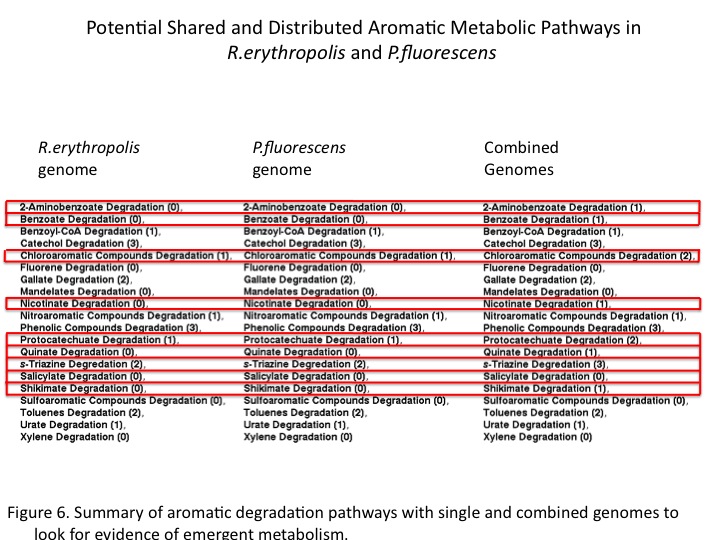
| + | Finally, the sums of general aromatic degradation pathways were compared for the organisms genomes separately and together (Figure 6). This resulted in emergent predicted pathways in combination as well as combinatorial increases araomatic degradation potential. Ultimately, a gene annotation based models for distributed metabolism in the environment may help to engineer and optimize complex metabolism through synthetic consortia. </br> | ||
| + | |||
| + | <p align=center><img src="https://static.igem.org/mediawiki/2012/a/aa/UbcigemSlide6.jpg"></p> | ||
| + | |||
| + | [1] Hanson, N.W; Page, A.P; Konwar, K.M; Howes, C.G; Hallam, S.J. Metabolic interaction networks for the whole community, 2012. Unpublished.</br></br> | ||
| + | |||
| + | [2] Kayser, K.J; Biolaga-Jones, B.A.; Jackowski, K; Odusan, O; Kildane, J.J. Utilzation of organosulfur compounds by anexic and mixed culture of Rhodococcus rhodochrous IGTS8, 1993. Journal of General Microbioology, 139: 3123-3129.</br></br> | ||
| + | |||
| + | [3] Goswami, M; Shivaraman, N; Singh, R.P. Microbial metabolism of 2-chlorophenol, phenol and p-cresol by Rhodococcus erythropolis M1 and co-culture with Pseudomonas fluorescens P1, 2005. Microbiological Research, 160: 101-109.</br></br> | ||
Revision as of 00:18, 4 October 2012

Pathway Model
The study of environmental genomics attempts to capture the taxonomic and functional diversity of natural microbial communities. Our host at UBC, the Hallam lab, designs novel tools for analyzing the gene content in the context of distributed metabolism. Recently, a pipeline has been developed for the automated construction and visualizing of metabolic pathways from genomic data by integrating software such as Pathway Tools, Pathologic and Metacyc [1]. This provided an opportunity to model pathway compartmentalizing and distribution between microbes in the natural environment as it applies to our project. Summary of the pipeline for community-level metabolic analysis (Figure 1): First, open reading frames predicted from sequence data using Prodigal are annotated by protein BLAST and summarized in a GenBank file. Pathway/genome databases (PGDBs) are generated from sequence data in a manner that does not constrain predictions within the scope of model organisms. This allows for a community-based analysis that can be visualized using Pathway Tools.




 "
"