Team:Tokyo Tech/Projects/rose pattern/index.htm
From 2012.igem.org
(→Project Overview) |
(→Cell-Cell communication Project OVERVIEW) |
||
| (65 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{tokyotechcss}} | {{tokyotechcss}} | ||
| - | |||
| - | |||
| - | |||
{{tokyotechmenubar}} | {{tokyotechmenubar}} | ||
<div class="whitebox"> | <div class="whitebox"> | ||
| + | <br><br><br><br> | ||
<div id="tokyotech" style=" font:bold ;left ; font-size: 50px; color: #1E90FF; padding: 10px;"> | <div id="tokyotech" style=" font:bold ;left ; font-size: 50px; color: #1E90FF; padding: 10px;"> | ||
| - | + | Template </div> | |
</div class="whitebox"> | </div class="whitebox"> | ||
<div id="tokyotech" style=" font:bold ;left ; font-size: 15px; color: #000000; padding: 10px;"> | <div id="tokyotech" style=" font:bold ;left ; font-size: 15px; color: #000000; padding: 10px;"> | ||
| - | |||
<div class="whitebox"> | <div class="whitebox"> | ||
[[File:tokyotechmakerose.png|200px|right]] | [[File:tokyotechmakerose.png|200px|right]] | ||
| + | __TOC__ | ||
| + | |||
| + | =Abastract= | ||
| + | [[#References|[1]]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =References= | ||
| + | '''1.''' | ||
| + | |||
=Achievement= | =Achievement= | ||
| - | |||
| - | |||
| - | |||
| - | |||
TEXT | TEXT | ||
| + | [[File:tokyotechrommejulistory.gif|right]] | ||
| + | |||
| + | |||
| + | =Links= | ||
| + | Tokyo tech parts | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934011 BBa_K934011] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934012 BBa_K934012] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934013 BBa_K934013] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934016 BBa_K934016] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934022 BBa_K934022] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934023 BBa_K934023] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934024 BBa_K934024] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934025 BBa_K934025] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934026 BBa_K934026] | ||
| + | |||
| + | |||
| + | ==CEll-cell other collage== | ||
| + | ===3OC6HSLdependent3OC12HSLproduction=== | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K081016 BBa_K081016] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_R0062 BBa_R0062] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===Positivefeedbackassay=== | ||
| + | Text | ||
| + | |||
| + | ==PHA other collage== | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K125801 BBa_K125801] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K125802 BBa_K125802] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K125803 BBa_K125803] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K125804 BBa_K125804] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K125501 BBa_K125501] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K125502 BBa_K125502] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K125503 BBa_K125503] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K125504 BBa_K125504] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156018 BBa_K156018] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K282000 BBa_K282000] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K338003 BBa_K338003] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K338004 BBa_K338004] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156022 BBa_K156022] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156031 BBa_K156031] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156033 BBa_K156033] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156034 BBa_K156034] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156012 BBa_K156012] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156013 BBa_K156013] | ||
| + | |||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156014 BBa_K156014] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156021 BBa_K156021] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156018 BBa_K156018] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156019 BBa_K156019] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156020 BBa_K156020] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156022 BBa_K156022] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K156023 BBa_K156023] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K342001 BBa_K342001] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K934026 BBa_K934026] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K089001 BBa_K089001] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K089002 BBa_K089002] | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K089003 BBa_K089003] | ||
| + | |||
| + | [https://2008.igem.org/Team:Hawaii iGEM08_Hawaii] | ||
| + | |||
| + | [https://2008.igem.org/Team:Virginia iGEM08_Virginia] | ||
| + | |||
| + | [https://2008.igem.org/Team:tsinghua iGEM08_tsinghua] | ||
| + | |||
| + | [https://2008.igem.org/Team:Utah State iGEM08_Utah State] | ||
| + | |||
| + | [https://2009.igem.org/Team:Duke iGEM09_Duke] | ||
| + | |||
| + | [https://2010.igem.org/Team:Caltech iGEM10_Caltech] | ||
| + | |||
| + | [https://2010.igem.org/Team:INSA-Lyon iGEM10_INSA-Lyon] | ||
| + | aaaa | ||
| + | |||
| + | =Introduction= | ||
==What's PHA== | ==What's PHA== | ||
| + | [[File:tokyotech PHA Chemical synthesis of PHA .png|350px|thumb|right|fig1,synthetic mechanism]] | ||
| + | PHAs are the abbreviation of Polyhydroixyalkanoates, which are linear polyesters produced in nature by bacterial fermentation of sugar and lipids. They are produced by the bacteria to store carbon and energy. PHAs are also a kind of bio-plastics, which are more sustainable because they can be biodegraded in the environment a lot more faster than fossil-fuel plastics. PHBs are the abbreviation of polyhydrobutyrates ,which are a kind of polyhydroalkanoates. In our project we used the poly-3-hydroxybutyrate(P3HB)form of PHBs, which is the most common type of polyhydroxyalkanoate synthesized by the gene phaC1-A-B from Ralstonia eutropha H16. | ||
| + | |||
| + | As long as we know ,there was no team succeeded in synthesizing PHAs in iGEM format before. But this year we are very excited to tell you that we made it and we are going to show it to all of you. | ||
| + | 9/4 | ||
| + | |||
| + | =Project Overview= | ||
| + | <div class="whitebox"> | ||
| + | ===Pre-experiment=== | ||
| + | |||
| + | Before the igem format vector coming,we actually synthesized PHA by using the vector PSB1C3 on E.coli JM109.We synthesized PHA by following steps. | ||
| + | 1.Take the right gene phaC1-A-B from R.eutropha, and increase it by PCR | ||
| + | 2.Insert the gene into the PSB1C3 vector by using a series of genetic operation including ligation and ethanol precipitation | ||
| + | 3. Put the plasmid into E.coli JM109 by using the technique of transformation and plant it on LBmedium with Cm and Nile-Red then culture it overnight. | ||
| + | 4. Check whether colonies are stained orange under UV | ||
| + | 5. Produce PHAs by quantity by main culture the E.coli | ||
| + | 6. Collect the strains and freeze dry it. | ||
| + | 7. Stain the product and Measure the weight | ||
| + | |||
| + | Assay | ||
| + | In order to check whether the product is PHA or not ,we used several ways of assay. | ||
| + | 1. Read the gene sequence of the gene | ||
| + | 2. See whether the product can be stained by Nile-Red or Nile-Blue | ||
| + | 3. Make sure the product under microscope | ||
| + | 9/4 | ||
| + | </div> | ||
| + | <div class="whitebox"> | ||
| + | =Production of PHB main culture= | ||
| + | ==Protocol== | ||
| + | 1 Wash Erlenmeyer flasks with mild detergent then rinse with distilled water. | ||
| + | |||
| + | 2 Measure 4% LB medium and add it to each Erlenmeyer flask inside clean bench. | ||
| + | |||
| + | 3 Add 96% distilled water to each Erlenmeyer flask and cover the flasks with four-folded aluminum foil. | ||
| + | |||
| + | 4 Set all flasks into autoclave | ||
| + | |||
| + | 5 Clear the clean bench | ||
| + | |||
| + | 6 Add 1/1000 Chloramphenicol and 4% Glucose solution (50%) after the medium is completely cooled. | ||
| + | |||
| + | 7 Add 2% preculture solution into each flasks and shaking culture at 37 ° C for 72 hours. | ||
| + | |||
| + | =====Collection of PHBs in JM109===== | ||
| + | 1 Weigh empty 50ml falcon tube without lid and make a record. | ||
| + | |||
| + | 2 Add some culture solution into each tube. | ||
| + | |||
| + | 3 Set the tubes into centrifuge and make sure that the label faces outside. | ||
| + | |||
| + | 4 4 ° C ,5000G ,10min in centrifuge. | ||
| + | |||
| + | 5 Remove the supernatant with electric pipettor then add culture solution and set in centrifuge again. | ||
| + | |||
| + | 6 After adding all the culture solution and setting in centrifuge, remove the supernatant and add water, set in centrifuge again. | ||
| + | |||
| + | 7 Remove the supernatant and add a little amount of water. | ||
| + | |||
| + | 8 Cover the tubes with double layers of parafilms and fully freeze them. | ||
| + | |||
| + | Freeze drying | ||
| + | |||
| + | 1 Poke several holes on the tubes’ parafilm with toothpick. | ||
| + | |||
| + | 2 Set the tubes on the freeze drying machine. | ||
| + | |||
| + | 3 Freeze dry for 3 days. | ||
| + | |||
| + | date9/4 | ||
| + | </div> | ||
| + | |||
| + | |||
| + | {| class="wikitable" cellpadding="5" | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | |- | ||
| + | | Colorblindness||content||content|| style="background:#92D050" |kore'''kaigyou''' || style="background:#92D050" | kaigyou '''kore'''||content.#######||content.####### | ||
| + | |- | ||
| + | | Familial Hypercholesterolemia||content||content||style="background:#92D050" |content||style="background:#92D050" | content||content.#######||content.####### | ||
| + | |- | ||
| + | | Myc-gene Cancer||Text||Text||Text||style="background:#92D050" |Text ||Text||Text.####### | ||
| + | |- | ||
| + | | Text||Text||Text||Text||style="background:#92D050" |Text ||Text||Text.####### | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| class="wikitable" cellpadding="3" | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''Text''' | ||
| + | |- | ||
| + | | Colorblindness||content||content|| style="background:#92D050" |kore'''kaigyou''' || style="background:#92D050" | kaigyou '''kore'''||content.#######||content.####### | ||
| + | |- | ||
| + | | Familial Hypercholesterolemia||content||content||style="background:#92D050" |content||style="background:#92D050" | content||content.#######||content.####### | ||
| + | |- | ||
| + | | Myc-gene Cancer||Text||Text||Text||style="background:#92D050" |Text ||Text||Text.####### | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| style="width:500px;align:right;background:#f5f5f5;text-align:right;font-family: helvetica, arial, sans-serif;color:#555555;margin-top:5px;" cellspacing="20" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:2em;color:#ea8828;"|Gene Circuit | ||
| + | |align="right"|<html><iframe width="480" height="360" src="http://www.youtube.com/embed/kDwqsIgsoQ4" frameborder="0" allowfullscreen></iframe></html> | ||
| + | |||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | {| style="font-family:'Andale Mono','Times New Roman'" border="1" | ||
| + | | align="center" style="background:#f0f0f0;"|'''TextA''' | ||
| + | | align="center" style="background:#f0f0f0;"|'''TextA''' | ||
| + | |- | ||
| + | | TextB||TextB | ||
| + | |- | ||
| + | | TextB||TextB | ||
| + | |- | ||
| + | | TextB||TextB | ||
| + | |- | ||
| + | | TextB||TextB | ||
| + | |- | ||
| + | | TextB||TextB | ||
| + | |- | ||
| + | | TextB||TextBv | ||
| + | |- | ||
| + | | TextB||TextB|- | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | {{tokyotechcss}} | ||
| + | {{tokyotechtop}} | ||
| + | {{tokyotechmenubar}} | ||
| + | <div class="whitebox"> | ||
| + | <div id="tokyotech" style=" font:bold ;left ; font-size: 50px; color: #1E90FF; padding: 10px;"> | ||
| + | cell-cell communication </div> | ||
| + | </div class="whitebox"> | ||
| + | <div class="whitebox"> | ||
| + | <div id="tokyotech" style=" font:Arial ;left ; font-size: 15px; color: #000000; padding: 30px;"> | ||
| + | __TOC__ | ||
| + | =Scene1 Fall in love Positive feedback mechanism= | ||
| + | |||
| + | ==LasR generator== | ||
| + | [[File:gene circuit positive feedback tokyotech LasR assaydeta.png|450px|thumb|left|fig5,LasR generator]] | ||
| + | [[File:gene circuit positive feedback tokyotech LasR assay.png|300px|thumb|right|fig5,LasR generator]] | ||
| + | [[File:gene circuit positive feedback tokyotech LasR assayparts.png|350px|thumb|right|fig5,LasR generator deta]] | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | Text...[[ Team:Tokyo_Tech/Projects/LasR generator/index.htm | see more]] | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
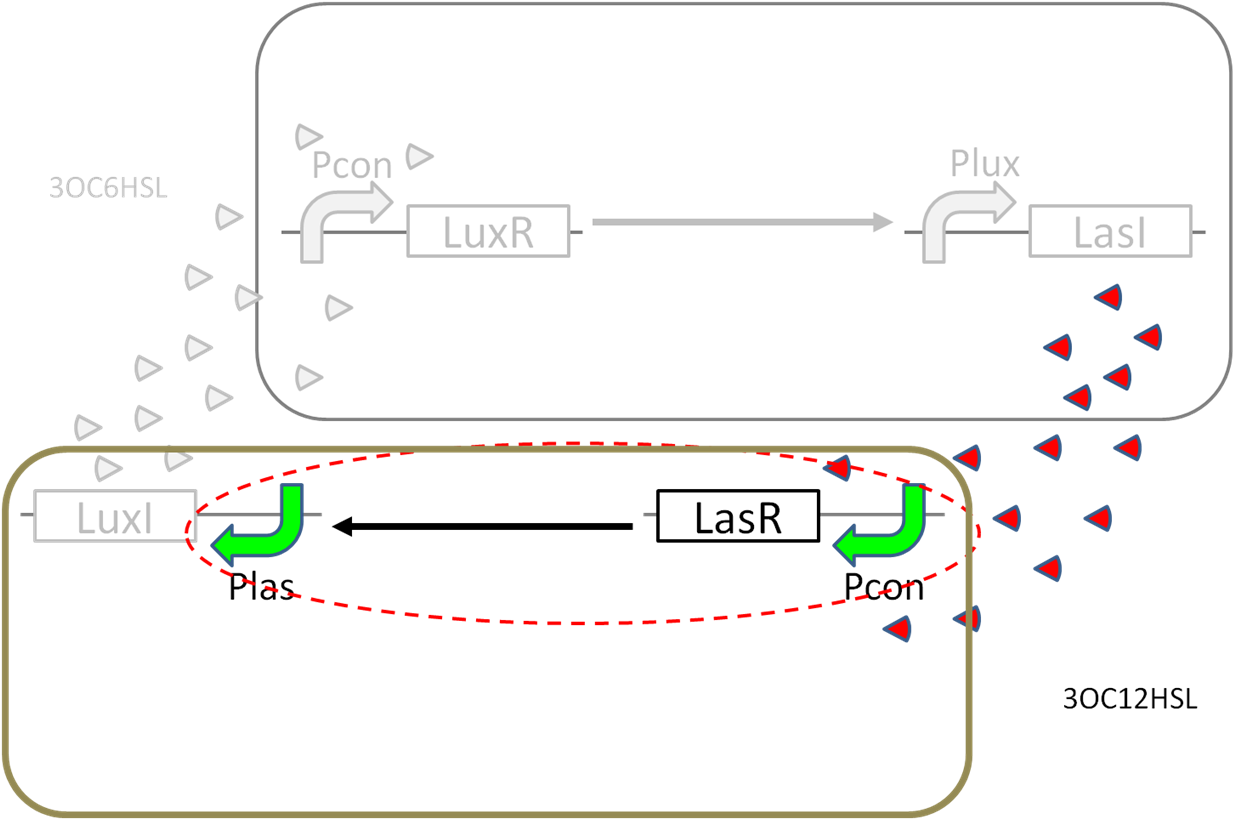
| + | ==3OC6HSL→pLuxLasI-PLas== | ||
| + | [[File:gene circuit positive feedback tokyotech 3OC6HSL→pLuxLasI-PLas assaydeta.png|450px|thumb|left|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | [[File:gene circuit positive feedback tokyotech 3OC6HSL→pLuxLasI-PLas assay.png|300px|thumb|right|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | [[File:gene circuit positive feedback tokyotech 3OC6HSL→pLuxLasI-PLas assayparts.png|330px|thumb|right|fig5,3OC6HSL→pLuxLasI-PLas deta]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | Text...[[ Team:Tokyo_Tech/Projects/3OC6HSL→pLuxLasI-PLas/index.htm | see more]] | ||
| + | <br><br><br><br><br><br><br> | ||
| + | |||
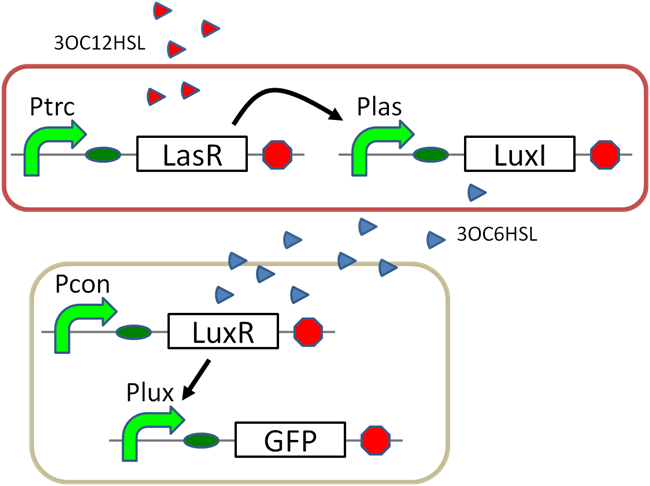
| + | ==3OC12HSL→pLasLuXI-PLux== | ||
| + | [[File:gene circuit positive feedback tokyotech 3OC12HSL→pLasLuXI-PLux assayparts.png|450px|thumb|left|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | [[File:gene circuit positive feedback tokyotech 3OC12HSL→pLasLuXI-PLux assay.png|300px|thumb|right|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | [[File:gene circuit positive feedback tokyotech 3OC12HSL→pLasLuXI-PLux assaydeta.png|320px|thumb|right|fig5,3OC6HSL→pLuxLasI-PLas deta]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | Text...[[ Team:Tokyo_Tech/Projects/3OC12HSL→pLasLuxI-PLux/index.htm | see more]] | ||
| + | <br><br><br><br><br><br> | ||
| + | |||
| + | ==positive feedback assay== | ||
| + | [[File:gene circuit positive feedback tokyotech positive feedback assayparts.png|450px|thumb|left|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | [[File:gene circuit positive feedback tokyotech positive feedback assay.png|300px|thumb|right|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | [[File:gene circuit positive feedback tokyotech positive feedback assaydeta.png|320px|thumb|right|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | =Band detect system= | ||
| + | |||
| + | ==Lux-Tet hybrid promoter assay== | ||
| + | [[File:gene circuit band detect system tokyotech Lux-Tet hybrid promoter assay parts.png|450px|thumb|left|fig5,Lux-Tet hybrid promoter assay]] | ||
| + | [[File:gene circuit band detect system tokyotech Lux-Tet hybrid promoter assay.png|300px|thumb|right|fig5,Lux-Tet hybrid promoter assay]] | ||
| + | [[File:gene circuit band detect system tokyotech Lux-Tet hybrid promoter assaydeta.png|320px|thumb|right|fig5,Lux-Tet hybrid promoter assay deta]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | Text...[[ Team:Tokyo_Tech/Projects/Lux-Tet hybrid promoter/index.htm | see more]] | ||
| + | <br><br><br><br><br><br><br><br><br> | ||
| + | |||
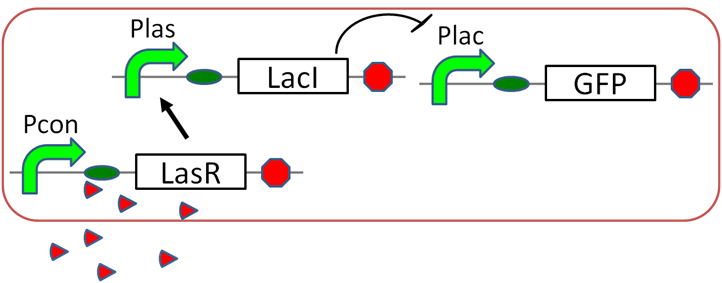
| + | =Scene3 Romeo Suicide = | ||
| + | |||
| + | ==Las→Lac assay== | ||
| + | [[File:gene circuit Suicide mechanism tokyotech Las→Lac assay, Lux→Lac assayparts.png|300px|thumb|left|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | [[File:gene circuit Suicide mechanism tokyotech Las→Lac assay, Lux→Lac assaydeta.png|250px|thumb|left|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br> | ||
| + | |||
| + | =Scene4 Juliet Suicide= | ||
| + | ==Lux→Lac assay== | ||
| + | [[File:gene circuit Suicide mechanism tokyotech Las→Lac assay, Lux→Lac assayparts2.png|300px|thumb|left|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | [[File:gene circuit Suicide mechanism tokyotech Las→Lac assay, Lux→Lac assaydeta2.png|250px|thumb|left|fig5,3OC6HSL→pLuxLasI-PLas]] | ||
| + | <br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | Text...[[ Team:Tokyo_Tech/Projects/Las→Lac assay, Lux→Lac assay/index.htm | see more]] | ||
| + | |||
| + | =modeling= | ||
| + | [[File:tokyotechromijurimodeling.png|350px|thumb|right|Romeo&Juliette signal change with times]] | ||
| + | |||
Text | Text | ||
| + | <br><br><br><br><br><br><br><br>><br><br> | ||
| + | [[File:tokyotechromijurimodeling formula.png|850px|thumb|center|Romeo&Juliette Formula]] | ||
| - | = | + | =Conclusion= |
| - | + | We succeeded in constructing (number) new BioBrick, (name). We characterized mechanism. From the experimental data and simulation result, we confirmed the feasibility of the project "Romeo and Juliette". | |
| - | + | ||
| - | + | ||
Latest revision as of 20:44, 22 September 2012
Contents |
Abastract
References
1.
Achievement
TEXT
Links
Tokyo tech parts
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934001 BBa_K934001]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934011 BBa_K934011]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934012 BBa_K934012]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934013 BBa_K934013]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934016 BBa_K934016]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934022 BBa_K934022]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934023 BBa_K934023]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934024 BBa_K934024]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934025 BBa_K934025]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934026 BBa_K934026]
CEll-cell other collage
3OC6HSLdependent3OC12HSLproduction
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K081016 BBa_K081016]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_R0062 BBa_R0062]
Positivefeedbackassay
Text
PHA other collage
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K125801 BBa_K125801]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K125802 BBa_K125802]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K125803 BBa_K125803]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K125804 BBa_K125804]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K125501 BBa_K125501]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K125502 BBa_K125502]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K125503 BBa_K125503]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K125504 BBa_K125504]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156018 BBa_K156018]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K282000 BBa_K282000]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K338003 BBa_K338003]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K338004 BBa_K338004]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156022 BBa_K156022]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156031 BBa_K156031]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156033 BBa_K156033]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156034 BBa_K156034]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156012 BBa_K156012]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156013 BBa_K156013]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156014 BBa_K156014]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156021 BBa_K156021]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156018 BBa_K156018]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156019 BBa_K156019]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156020 BBa_K156020]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156022 BBa_K156022]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K156023 BBa_K156023]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K342001 BBa_K342001]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K934026 BBa_K934026]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K089001 BBa_K089001]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K089002 BBa_K089002]
[http://partsregistry.org/wiki/index.php?title=Part:BBa_K089003 BBa_K089003]
iGEM10_INSA-Lyon aaaa
Introduction
What's PHA
PHAs are the abbreviation of Polyhydroixyalkanoates, which are linear polyesters produced in nature by bacterial fermentation of sugar and lipids. They are produced by the bacteria to store carbon and energy. PHAs are also a kind of bio-plastics, which are more sustainable because they can be biodegraded in the environment a lot more faster than fossil-fuel plastics. PHBs are the abbreviation of polyhydrobutyrates ,which are a kind of polyhydroalkanoates. In our project we used the poly-3-hydroxybutyrate(P3HB)form of PHBs, which is the most common type of polyhydroxyalkanoate synthesized by the gene phaC1-A-B from Ralstonia eutropha H16.
As long as we know ,there was no team succeeded in synthesizing PHAs in iGEM format before. But this year we are very excited to tell you that we made it and we are going to show it to all of you. 9/4
Project Overview
Pre-experiment
Before the igem format vector coming,we actually synthesized PHA by using the vector PSB1C3 on E.coli JM109.We synthesized PHA by following steps. 1.Take the right gene phaC1-A-B from R.eutropha, and increase it by PCR 2.Insert the gene into the PSB1C3 vector by using a series of genetic operation including ligation and ethanol precipitation 3. Put the plasmid into E.coli JM109 by using the technique of transformation and plant it on LBmedium with Cm and Nile-Red then culture it overnight. 4. Check whether colonies are stained orange under UV 5. Produce PHAs by quantity by main culture the E.coli 6. Collect the strains and freeze dry it. 7. Stain the product and Measure the weight
Assay In order to check whether the product is PHA or not ,we used several ways of assay. 1. Read the gene sequence of the gene 2. See whether the product can be stained by Nile-Red or Nile-Blue 3. Make sure the product under microscope 9/4
Production of PHB main culture
Protocol
1 Wash Erlenmeyer flasks with mild detergent then rinse with distilled water.
2 Measure 4% LB medium and add it to each Erlenmeyer flask inside clean bench.
3 Add 96% distilled water to each Erlenmeyer flask and cover the flasks with four-folded aluminum foil.
4 Set all flasks into autoclave
5 Clear the clean bench
6 Add 1/1000 Chloramphenicol and 4% Glucose solution (50%) after the medium is completely cooled.
7 Add 2% preculture solution into each flasks and shaking culture at 37 ° C for 72 hours.
Collection of PHBs in JM109
1 Weigh empty 50ml falcon tube without lid and make a record.
2 Add some culture solution into each tube.
3 Set the tubes into centrifuge and make sure that the label faces outside.
4 4 ° C ,5000G ,10min in centrifuge.
5 Remove the supernatant with electric pipettor then add culture solution and set in centrifuge again.
6 After adding all the culture solution and setting in centrifuge, remove the supernatant and add water, set in centrifuge again.
7 Remove the supernatant and add a little amount of water.
8 Cover the tubes with double layers of parafilms and fully freeze them.
Freeze drying
1 Poke several holes on the tubes’ parafilm with toothpick.
2 Set the tubes on the freeze drying machine.
3 Freeze dry for 3 days.
date9/4
| Text | Text | Text | Text | Text | Text | Text |
| Colorblindness | content | content | korekaigyou | kaigyou kore | content.####### | content.####### |
| Familial Hypercholesterolemia | content | content | content | content | content.####### | content.####### |
| Myc-gene Cancer | Text | Text | Text | Text | Text | Text.####### |
| Text | Text | Text | Text | Text | Text | Text.####### |
| Text | Text | Text | Text | Text | Text | Text |
| Colorblindness | content | content | korekaigyou | kaigyou kore | content.####### | content.####### |
| Familial Hypercholesterolemia | content | content | content | content | content.####### | content.####### |
| Myc-gene Cancer | Text | Text | Text | Text | Text | Text.####### |
| Gene Circuit |
| TextA | TextA |
| TextB | TextB |
| TextB | TextB |
| TextB | TextB |
| TextB | TextB |
| TextB | TextB |
| TextB | TextBv |
| TextB | - |
Scene1 Fall in love Positive feedback mechanism
LasR generator
Text... see more
3OC6HSL→pLuxLasI-PLas
Text... see more
3OC12HSL→pLasLuXI-PLux
Text... see more
positive feedback assay
Band detect system
Lux-Tet hybrid promoter assay
Text... see more
Scene3 Romeo Suicide
Las→Lac assay
Scene4 Juliet Suicide
Lux→Lac assay
Text... see more
modeling
Text
>
 "
"