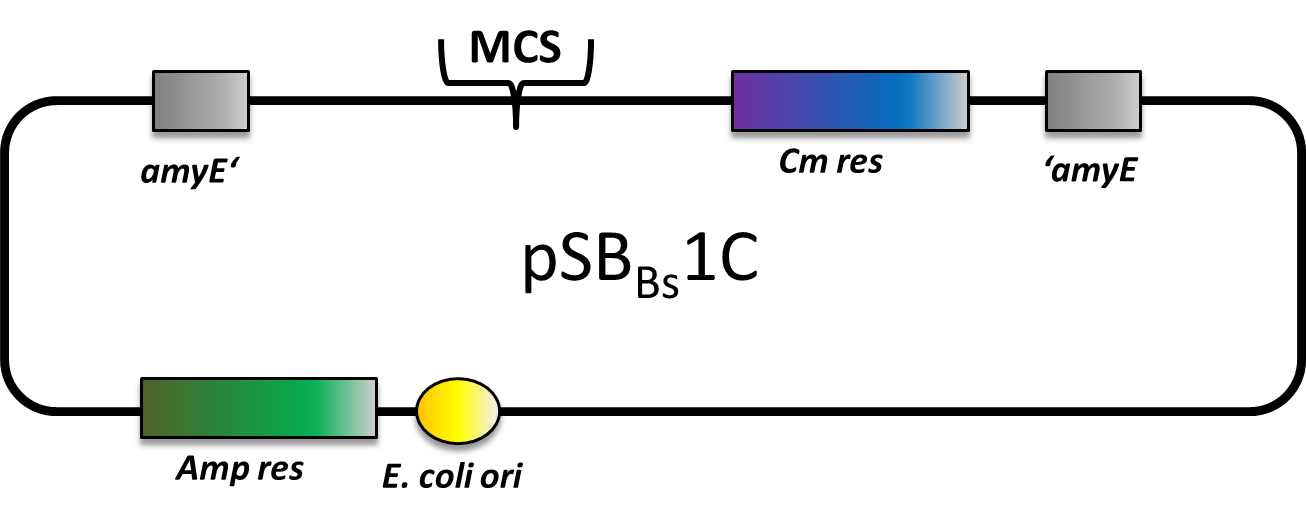Team:LMU-Munich/Bacillus BioBricks
From 2012.igem.org
| (322 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<!-- Include the next line at the beginning of every page --> | <!-- Include the next line at the beginning of every page --> | ||
{{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_culture_tubes.resized.jpg|3}} | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_culture_tubes.resized.jpg|3}} | ||
| - | + | [[File:Bacillus BioBrick Box banner.resized WORDS.JPG|620px|link=]] | |
| - | [[ | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | [[Image:BacillusBioBrickBox.png|100px|right|link=]] | ||
| + | =='''B<sup>4</sup>''' - 22 core parts for ''Bacillus subtilis''== | ||
| + | <br> | ||
| + | <p align="justify">A major goal of our iGEM project is to [https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction introduce ''B. subtilis''] as a new chassis for BioBrick-based synthetic biology. For that purpose, we created a toolbox of <i>Bacillus</i> BioBricks to contribute to the registry to make it accessible to many more future iGEM-teams and the entire public microbiology domain! This ''<b>Bacillus'' B</b>io<b>B</b>rick <b>B</b>ox ('''B<sup>4</sup>''') contains the following ''Bacillus'' specific parts:</p> | ||
| - | |||
| - | |||
| - | + | <div class="box"> | |
| + | {| width="100%" | ||
| - | + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks/Vectors|Vectors]]''' | |
| - | |- | + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks/Promoters|Promoters]]''' |
| - | + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks/Reporters|Reporters]]''' | |
| - | + | | style="width:20%"|'''[[Team:LMU-Munich/Bacillus_BioBricks/Tags|Affinity tags]] | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | | | + | |
|- | |- | ||
| - | | | + | |[[File:LMU Backbone.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors|Vectors]] |
| - | | | + | |[[File:LMU PromoterIconBC.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Promoters]] |
| - | | | + | |[[File:LMU Reporter.png|60px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Reporters]] |
| - | | | + | |[[File:Proteinaffinitytagbutton.png|50px|link=Team:LMU-Munich/Bacillus_BioBricks#Affinity_Tags]] |
| - | | | + | |
| - | | | + | |
| - | | | + | |
|- | |- | ||
| - | |< | + | |valign="top"|<font size="2"> |
| - | + | pSB<sub>''Bs''</sub>1C<br> | |
| - | + | pSB<sub>''Bs''</sub>4S<br> | |
| - | + | pSB<sub>''Bs''</sub>2E<br> | |
| - | + | pSB<sub>''Bs''</sub>1C-''lac''Z<br> | |
| - | + | pSB<sub>''Bs''</sub>3C-''lux''ABCDE<br> | |
| - | + | pSB<sub>''Bs''</sub>4S-P<sub>''xyl''</sub><br> | |
| - | + | pSB<sub>''Bs''</sub>0K-P<sub>''spac''</sub><br> | |
| - | + | '''Sporo'''vector</font> | |
| - | + | |valign="top"|<font size="2"> | |
| - | + | Anderson<br> | |
| - | + | P<sub>''liaG''</sub><br> | |
| - | + | P<sub>''veg''</sub><br> | |
| - | + | P<sub>''lepA''</sub><br> | |
| - | + | P<sub>''liaI''</sub><br> | |
| - | + | ''xylR''-P<sub>''xyl''</sub></font> | |
| - | + | |valign="top"|<font size="2"> | |
| - | + | ''gfp''<br> | |
| - | + | ''mkate2''<br> | |
| - | + | ''lacZ''<br> | |
| - | + | ''luc+''<br></font> | |
| - | + | |valign="top"|<font size="2"> | |
| - | + | Flag<br>His<br>cMyc<br>Strep<br>HA</font> | |
| - | + | ||
| - | + | ||
| - | | | + | |
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | |< | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
|} | |} | ||
| + | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |- | + | [[File:NEXT.png|right|80px|link=Team:LMU-Munich/Germination_Stop]] [[File:BACK.png|left|80px|link=Team:LMU-Munich/Data/differentiation_tour]] |
| - | | | + | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | = | + | <p align="justify">Since ''Bacillus subtilis'' is not an organism commonly used in iGEM, please check out our [https://2012.igem.org/Team:LMU-Munich/Bacillus_Introduction Introduction] to learn more about it.</p> |
| - | + | ||
| + | <div class="box"> | ||
| + | ==''Bacillus'' Vectors [[File:LMU Backbone.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks#Bacillus_Vectors|Vectors]]== | ||
| + | {| "width=100%" style="text-align:center;" style="align:right"| | ||
| + | |<p align="justify">We have generated a suite of BioBrick-compatible vectors: three empty insertional backbones with different antibiotic resistances and integration loci, two reporter and two expression vectors.</p> | ||
| + | |[[File:LMU-Munich-PSBBs1C.png|200px|right|link=Team:LMU-Munich/Bacillus BioBricks/Vectors]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Bacillus BioBricks/Vectors]] | ||
| + | |} | ||
| + | </div> | ||
| - | ==Bacillus | + | <div class="box"> |
| + | ==''Bacillus'' Promoters [[File:LMU PromoterIconBC.png|100px]]== | ||
| + | {| "width=100%" style="align:right"| | ||
| + | | | ||
| + | <p align="justify">To provide a set of promoters of different strength we characterized several promoters in ''Bacillus subtilis''. Both constitutive and inducible promoters are covered.</p> | ||
| + | | | ||
| + | [[File:Promoters overview.png|200px|right|link=Team:LMU-Munich/Bacillus BioBricks/Promoters]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Bacillus BioBricks/Promoters]] | ||
| + | |} | ||
| + | </div> | ||
| - | - | + | <div class="box"> |
| - | - | + | ==''Bacillus'' Reporters [[File:LMU Reporter.png|50px]]== |
| - | - | + | {| "width=100%" style="align:right"| |
| - | - | + | | |
| + | <p align="justify">We designed and codon-optimized a set of reporters that are commonly used in ''B. subtilis''.</p> | ||
| + | | | ||
| + | [[File:LMU GFP.jpg|200px|right|link=Team:LMU-Munich/Bacillus BioBricks/Reporters]] | ||
| + | |- | ||
| + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Bacillus BioBricks/Reporters]] | ||
| + | |} | ||
| + | </div> | ||
| - | + | <div class="box"> | |
| - | + | ==Affinity Tags [[File:Proteinaffinitytagbutton.png|50px]]== | |
| - | + | {| "width=100%" style="align:right"| | |
| - | + | | | |
| - | | | + | <p align="justify">We synthesized 5 affinity tags for protein purification. They all are designed in Freiburg standard with an optimized ribosome binding site upstream. We have not yet tested our tags.</p> |
| - | | | + | |
| - | |< | + | |
|- | |- | ||
| - | + | ! colspan="2" |[[File:LMU Arrow purple.png|40px|link=Team:LMU-Munich/Bacillus BioBricks/Tags]] | |
| - | + | |} | |
| - | + | </div> | |
| - | < | + | <br> |
| - | + | <br> | |
| - | + | <br> | |
| - | + | <br> | |
| - | <br | + | <div class="box"> |
| - | + | ====Project Navigation==== | |
| - | + | {| width="100%" align="center" style="text-align:center;" | |
| - | + | |[[File:Bacilluss_Intro.png|100px|link=Team:LMU-Munich/Bacillus_Introduction]] | |
| - | + | |[[File:BacillusBioBrickBox.png|100px|link=Team:LMU-Munich/Bacillus_BioBricks]] | |
| - | + | |[[File:SporeCoat.png|100px|link=Team:LMU-Munich/Spore_Coat_Proteins]] | |
| - | + | |[[File:GerminationSTOP.png|100px|link=Team:LMU-Munich/Germination_Stop]] | |
| - | + | |- | |
| - | + | |[[Team:LMU-Munich/Bacillus_Introduction|<font size="2">'''''Bacillus'''''<BR>Intro</font>]] | |
| - | + | |[[Team:LMU-Munich/Bacillus_BioBricks|<font size="2" face="verdana">'''''Bacillus'''''<BR>'''B'''io'''B'''rick'''B'''ox</font>]] | |
| - | + | |[[Team:LMU-Munich/Spore_Coat_Proteins|<font size="2" face="verdana">'''Sporo'''beads</font>]] | |
| - | + | |[[Team:LMU-Munich/Germination_Stop|<font size="2" face="verdana">'''Germination'''<BR>STOP</font>]] | |
| - | + | ||
| - | + | ||
| - | | | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | | | + | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
| + | </div> | ||
<!-- Include the next line at the end of every page --> | <!-- Include the next line at the end of every page --> | ||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Latest revision as of 16:41, 26 October 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]


B4 - 22 core parts for Bacillus subtilis
A major goal of our iGEM project is to introduce B. subtilis as a new chassis for BioBrick-based synthetic biology. For that purpose, we created a toolbox of Bacillus BioBricks to contribute to the registry to make it accessible to many more future iGEM-teams and the entire public microbiology domain! This Bacillus BioBrick Box (B4) contains the following Bacillus specific parts:
| Vectors | Promoters | Reporters | Affinity tags |

| 
| 
| |
|
pSBBs1C |
Anderson |
gfp |
Flag |
Since Bacillus subtilis is not an organism commonly used in iGEM, please check out our Introduction to learn more about it.
Bacillus Vectors 
We have generated a suite of BioBrick-compatible vectors: three empty insertional backbones with different antibiotic resistances and integration loci, two reporter and two expression vectors. | |
Bacillus Promoters 
|
To provide a set of promoters of different strength we characterized several promoters in Bacillus subtilis. Both constitutive and inducible promoters are covered. | |
Bacillus Reporters 
|
We designed and codon-optimized a set of reporters that are commonly used in B. subtilis. | |
Affinity Tags 
|
We synthesized 5 affinity tags for protein purification. They all are designed in Freiburg standard with an optimized ribosome binding site upstream. We have not yet tested our tags. | |

| 
| 
| 
|
| Bacillus Intro | Bacillus BioBrickBox | Sporobeads | Germination STOP |
 "
"