Team:NTNU Trondheim/Project
From 2012.igem.org
m (→Overall project - BACK: "Bacterial Anti-Cancer Kamikaze") |
(→Genes) |
||
| (22 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:NTNU_Trondheim/Templates/Header}} | {{:Team:NTNU_Trondheim/Templates/Header}} | ||
| + | <html> | ||
| + | <div class="container"> | ||
| + | <div class="page-header-top"> | ||
| + | <h1>Project description <small>Bacterial Anti-Cancer Kamikaze explained</small></h1> | ||
| + | </div> | ||
| + | </div> | ||
| - | |||
| + | <div class="container main-container"> | ||
| + | </html> | ||
__TOC__ | __TOC__ | ||
| - | == | + | ==Background== |
| - | + | Cancer is the leading cause of death worldwide. In 2008 alone, 13% of all deaths (7,6 million people) were caused by cancer, and it is not likely that the number will decrease. Lifestyle diseases, for instance, are huge risk factors for developing cancer, and diseases like type 2 diabetes are increasing rapidly on a worldwide basis. [http://www.who.int/gho/publications/world_health_statistics/2012/en/index.html], [http://www.who.int/mediacentre/factsheets/fs312/en/]. | |
| + | One of the most challenging aspects of cancer medicine is the difficulty of drug delivery directly at the tumor area. We therefore wanted to develop a system for specific drug delivery, to show that one of the most challenging medical problems can be solved by using synthetic biology. So we decided to make a system responding to factors specific for cancer cells, but without being too complicated [http://mct.aacrjournals.org/content/4/10/1636.full]. | ||
| + | In our system we want two different trigger factors, in order to lower the risk of releasing toxins in the healthy parts of the body. Our system is triggered by a low oxygen concentration and a high lactate concentration. When the ''E. coli'' detects these conditions, our system will activate the lysis genes, and the toxin will be released. | ||
| - | + | Differentiated cells (“normal” cells) can produce high amount of lactate in an anaerobic environment, but this is not a normal environment for the cells in our body. Under normal circumstances, the differentiated cells will primarily metabolize glucose to carbon dioxide in the citric acid cycle (TCA), and have a very low production of lactate. Cancer cells behave differently. They produce large amounts of lactate regardless of the oxygen concentration in their surroundings [http://www.ncbi.nlm.nih.gov/pubmed/19460998], [http://cancerres.aacrjournals.org/content/58/7/1408.full.pdf+html]. The reason that we want to use oxygen as a trigger factor is that tumors exists in an environment deprived of oxygen [http://www.springerlink.com/content/a60537w7085574v4/]. | |
| + | ==Overview== | ||
| - | [ | + | Our goal for this years competition is to create a genetic circuit which enables ''E. coli'' cells to detect and attack cancer cells. To do this, the cells should produce a tumor inhibitor or toxin effective at killing cancer cells, and respond to environmental cues indicating the presence of cancer cells by undergoing [http://en.wikipedia.org/wiki/Lysis lysis], releasing the anti-cancer agent into the surrounding environment. In principle, bacteria could be used in "search and destroy" missions against cancer inside the human body. We wish to develop our genetic circuit as a proof-of-concept of one such strategy. |
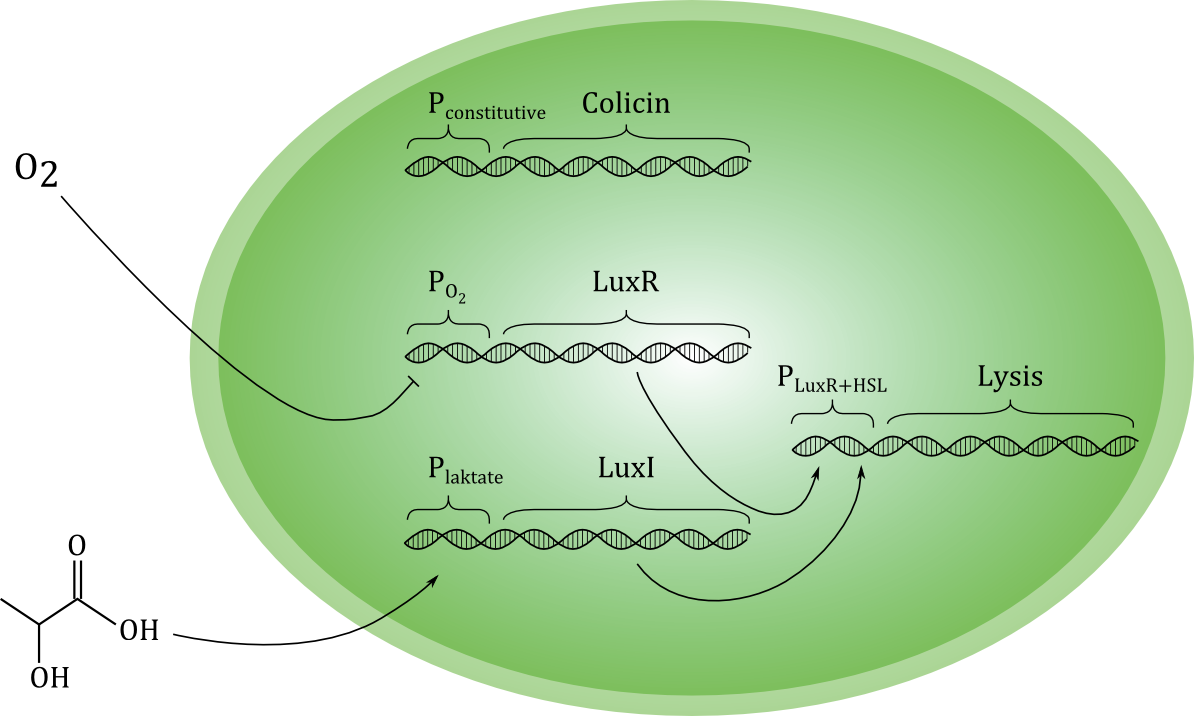
| + | A sketch of our current planned design for the circuit is shown below. | ||
| - | + | [[File:Grønn_krets.png|thumb|center|600px|A sketch of our planned genetic circuit]] | |
| - | + | Ideally, the toxin should only be released if cancer cells are actually encountered, and never otherwise. To achieve this, the lysis-inducing part of the circuit should be regulated such that it is highly unlikely to be activated in other situations. One possible way is to use a signal molecule that is solely associated with cancer cells to activate the lysis device. Another possibility is to use a combination of environmental cues that together indicate a high likelihood of cancer presence. We have chosen the latter strategy, and the environmental cues we aim to use are low O<sub>2</sub> and high lactate concentrations. | |
| - | + | Each of the two cues activates a separate gene, which encode different proteins. Together, these two proteins should activate the unit responsible for lysis. Therefore, if only one of the cues is present, the cell does not lyse, and toxin is not released in high levels. (Some leakage of toxin must be expected even under normal conditions with no activation of the circuit). Selective activation in the presence of cancer cells would be crucial for the concept to be effective in a medical situation. | |
| - | == | + | As shown in the sketch, we plan to express the toxin-producing gene with a constitutive promoter. This means that the toxin production is always active. Ideally, the toxin should be maximally effective against cancer cells and minimally toxic to the toxin-producing bacterial cell. We have evaluated several candidate molecules and decided to use the protein [http://proteopedia.org/wiki/index.php/Colicin_E1 Colicin E1]. |
| + | |||
| + | ==Details== | ||
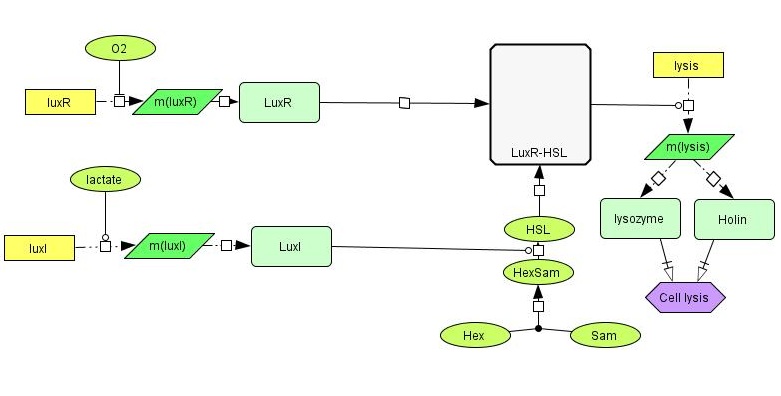
Below is a more detailed sketch of the circuit design ([https://2012.igem.org/File:NTNU_sketchlegend.jpg legend]). The environmental cues oxygen and lactate regulate the production of two regulatory proteins, LuxR and LuxI. Oxygen inhibits the production of LuxR, while lactate activates the production of LuxI. LuxI in turn catalyzes the formation of a homo-serine lactone (HSL) compound from S-Adenosyl Methionine (SAM) and Hexanoyl-ACP (Hex). When both LuxR and HSL is produced, they combine irreversibly to form a complex which activates the promoter in front of the lysis device (BioBrick part <partinfo>BBa_K112808</partinfo>, designed by the [https://2008.igem.org/Team:UC_Berkeley University of California Berkeley iGEM 2008] team). | Below is a more detailed sketch of the circuit design ([https://2012.igem.org/File:NTNU_sketchlegend.jpg legend]). The environmental cues oxygen and lactate regulate the production of two regulatory proteins, LuxR and LuxI. Oxygen inhibits the production of LuxR, while lactate activates the production of LuxI. LuxI in turn catalyzes the formation of a homo-serine lactone (HSL) compound from S-Adenosyl Methionine (SAM) and Hexanoyl-ACP (Hex). When both LuxR and HSL is produced, they combine irreversibly to form a complex which activates the promoter in front of the lysis device (BioBrick part <partinfo>BBa_K112808</partinfo>, designed by the [https://2008.igem.org/Team:UC_Berkeley University of California Berkeley iGEM 2008] team). | ||
| - | [[File:NTNU_sketch1b.jpg|center]] | + | [[File:NTNU_sketch1b.jpg|thumb|600px|center| Detailed overview of our genetic circuit.]] |
| - | In order to have the presence of lactate activate the production of LuxI, we need a lactate-sensitive [http://en.wikipedia.org/wiki/Promoter_%28genetics%29 promoter]. We | + | In order to have the presence of lactate activate the production of LuxI, we need a lactate-sensitive [http://en.wikipedia.org/wiki/Promoter_%28genetics%29 promoter]. We chose the lactate-sensitive promoter found in the [http://ecocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU164 lldPRD] operon of ''E. coli'' by PCR. However, the available literature indicates that this promoter [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC205257/ is repressed under low-oxygen conditions] due to the action of the ArcA regulatory protein. A possible solution is using site-directed mutagenesis to change the nucleotide sequence at the probable binding site of ArcA on the promoter to reduce its binding affinity and abolish its regulatory effect. |
==Challenges== | ==Challenges== | ||
| Line 33: | Line 46: | ||
First, how can cancer cells be reliably and effectively detected? Early in the process after deciding on the goal, we considered using several human signalling molecules such as HGF and VEGF as cancer indicators, as these are known to be over-produced by tumors. However, we were unable to determine quickly if these would be able to enter and affect ''E. coli'' cells, due to their large size. In contrast, O<sub>2</sub> and lactate are small molecules which are easily taken up by the cells, and are known to directly regulate the expression of various genes. | First, how can cancer cells be reliably and effectively detected? Early in the process after deciding on the goal, we considered using several human signalling molecules such as HGF and VEGF as cancer indicators, as these are known to be over-produced by tumors. However, we were unable to determine quickly if these would be able to enter and affect ''E. coli'' cells, due to their large size. In contrast, O<sub>2</sub> and lactate are small molecules which are easily taken up by the cells, and are known to directly regulate the expression of various genes. | ||
| - | == | + | ==Parts== |
| - | === | + | ===Promoters=== |
| - | To allow our system to work correctly, | + | To allow our system to work correctly, several different promoters are needed. Of these, the most simple is a constitutive (always on) promoter in front of the toxin-producing gene. For this purpose, we have extracted the BioBrick part <partinfo>BBa_J23119</partinfo> from the iGEM DNA distribution kit. Second, a promoter having higher activity at low oxygen levels, similar to those found in tumors. As a candidate for this function, the vgb promoter BioBrick part <partinfo>BBa_K561001</partinfo> was tested. Thirdly, a promoter that is activated by lactate, a possible indicator of tumor cells in the vicinity. We cloned the lldPp promoter of the ''E. coli'' [http://ecocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU164 lldPRD] operon for this, as well as the corresponding promoter from ''Corynebacterium glutamicum''. Lastly, a promoter that is activated by a compound resulting from the activation of the previous two units. In our sketch, this compound is a LuxR-HSL complex, which can activate the promoters of several BioBrick parts, such as <partinfo>BBa_K145150</partinfo> or <partinfo>BBa_R0062</partinfo>. |
| - | === | + | ===Genes=== |
LuxI is a acyl homoserine lactone syntase, an enzyme catalyzing the production of a homoserine lactone (HSL) from the precursors S-adenosyl methionine (Sam) and hexanoyl-ACP (Hex). The normal function of LuxI is as part of the bacterial lux system, a [http://en.wikipedia.org/wiki/Quorum_sensing quorom-sensing] system allowing cells in proximity to sense each others presence, estimate the local cell population and regulate [https://www.bio.cmu.edu/courses/03441/TermPapers/97TermPapers/lux/bioluminescence.html bioluminescence]. LuxI is available as the BioBrick part <partinfo>BBa_K092400</partinfo>. | LuxI is a acyl homoserine lactone syntase, an enzyme catalyzing the production of a homoserine lactone (HSL) from the precursors S-adenosyl methionine (Sam) and hexanoyl-ACP (Hex). The normal function of LuxI is as part of the bacterial lux system, a [http://en.wikipedia.org/wiki/Quorum_sensing quorom-sensing] system allowing cells in proximity to sense each others presence, estimate the local cell population and regulate [https://www.bio.cmu.edu/courses/03441/TermPapers/97TermPapers/lux/bioluminescence.html bioluminescence]. LuxI is available as the BioBrick part <partinfo>BBa_K092400</partinfo>. | ||
| - | [[File:NTNU_3OHSL.gif||center|[http://partsregistry.org/3OC6HSL 3-oxohexanoyl-homoserine lactone] is synthesized by LuxI]] | + | [[File:NTNU_3OHSL.gif|thumb|center|400px|[http://partsregistry.org/3OC6HSL 3-oxohexanoyl-homoserine lactone] is synthesized by LuxI]] |
LuxR is another protein in the lux system, and acts by binding to 3OC6HSL, the product formed by the catalytic activity of LuxI. LuxR-HSL then acts on gene promoters to regulate genetic expression. We will use the simultaneous expression of LuxR and LuxI leading to the the formation of LuxR-HSL to activate the lysis device which causes the cell to dissolve and release its produced toxin. | LuxR is another protein in the lux system, and acts by binding to 3OC6HSL, the product formed by the catalytic activity of LuxI. LuxR-HSL then acts on gene promoters to regulate genetic expression. We will use the simultaneous expression of LuxR and LuxI leading to the the formation of LuxR-HSL to activate the lysis device which causes the cell to dissolve and release its produced toxin. | ||
| - | The lysis device was made by the [https://2008.igem.org/Team:UC_Berkeley UC Berkeley iGEM 2008] team. The genes are from the [http://en.wikipedia.org/wiki/Enterobacteria_phage_T4 bacteriophage T4] (a virus that infects bacteria). We | + | The lysis device was made by the [https://2008.igem.org/Team:UC_Berkeley UC Berkeley iGEM 2008] team. The genes are from the [http://en.wikipedia.org/wiki/Enterobacteria_phage_T4 bacteriophage T4] (a virus that infects bacteria). We have placed the LuxR+HSL-activated promoter in front of the part to control the initiation of cell lysis. |
| - | + | ||
| - | + | ||
| + | [[File:S-Adenosyl methionine.png|thumb|center|300px|S-Adenoysyl-methionine (SAM)]] | ||
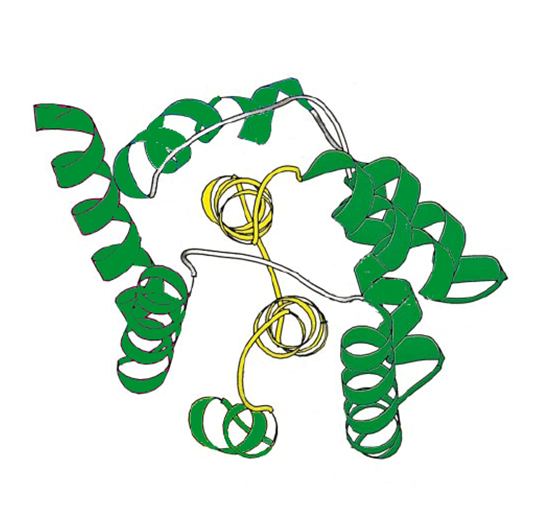
| - | + | Colicin E1 is a type of Colicin, a bacteriocin made by ''E. coli'',and which acts against other nearby ''E. coli'' by forming a pore in the membrane, leading to depolarisation of the membrane killing the cell [http://www.proteopedia.org/wiki/index.php/Colicin_E1]. Previous iGEM results indicate that this colicin may be effective against mammalian cancer cells as well [https://2008.igem.org/Team:Heidelberg/Project/Killing_II#Colicin-Receiver]. | |
| - | + | ||
| - | |||
| - | |||
| - | + | [[File:NTNU_ColicinE1_structure.png|thumb|300px|center|The structure of colicin E1.]] | |
| - | + | ||
| - | |||
| + | {{:Team:NTNU_Trondheim/Templates/Sponsors}}<html></div></div></html> | ||
| + | {{:Team:NTNU_Trondheim/Templates/Footer}} | ||
| + | <!--The structure of Hexanoyl-ACP: | ||
| + | [[file:HexanoylACP_structure.jpg|center|300px]] (from the RCSB Protein Databank, entry [http://www.rcsb.org/pdb/explore.do?structureId=2FAC 2FAC])--> | ||
| - | ==References== | + | <!--==References== |
Lactate sensitive promoter: | Lactate sensitive promoter: | ||
| Line 83: | Line 94: | ||
Lysis device: | Lysis device: | ||
| - | [https://2008.igem.org/Team:UC_Berkeley/LysisDevice UC Berkeley iGEM 2008: Lysis Device] | + | [https://2008.igem.org/Team:UC_Berkeley/LysisDevice UC Berkeley iGEM 2008: Lysis Device]--> |
Latest revision as of 20:43, 26 September 2012
 "
"