Team:Duke/Project
From 2012.igem.org
(Prototype team page) |
(→Summary of Theory/Introduction) |
||
| (40 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
<html> | <html> | ||
| - | < | + | |
| - | + | ||
| - | + | <style> | |
| - | + | /* Sidebar */ | |
| - | + | ||
| - | + | #sidebar { | |
| - | </ | + | float: right; |
| - | < | + | width: 253px; |
| - | + | } | |
| + | |||
| + | #sidebar ul { | ||
| + | margin: 0; | ||
| + | padding: 0; | ||
| + | list-style: none; | ||
| + | line-height: normal; | ||
| + | } | ||
| + | |||
| + | #sidebar li { | ||
| + | margin-bottom: 40px; | ||
| + | padding: 0 15px 0 20px; | ||
| + | } | ||
| + | |||
| + | #sidebar li ul { | ||
| + | } | ||
| + | |||
| + | #sidebar li li { | ||
| + | margin: 0; | ||
| + | padding: 5px 0; | ||
| + | border-bottom: 1px solid #E5E5E5; | ||
| + | } | ||
| + | |||
| + | #sidebar li h2 { | ||
| + | margin-bottom: 15px; | ||
| + | font-size: 18px; | ||
| + | } | ||
| + | |||
| + | |||
| + | /* Menu */ | ||
| + | |||
| + | #menu { | ||
| + | width: 960px; | ||
| + | height: 62px; | ||
| + | margin: 0; | ||
| + | border-top: 0; | ||
| + | border-right: 0; | ||
| + | border-left: 0; | ||
| + | padding: 0; | ||
| + | background: #252E3A url(http://i.imgur.com/bDFvK.jpg) no-repeat; | ||
| + | } | ||
| + | |||
| + | #menu ul { | ||
| + | margin: 0; | ||
| + | padding: 21px 0 0 30px; | ||
| + | list-style: none; | ||
| + | line-height: normal; | ||
| + | } | ||
| + | |||
| + | #menu li { | ||
| + | float: left; | ||
| + | padding: 0 20px 0 22px; | ||
| + | background: url(http://i.imgur.com/zJfPx.gif) no-repeat left center; | ||
| + | } | ||
| + | |||
| + | #menu li.first { | ||
| + | background: none; | ||
| + | } | ||
| + | |||
| + | #menu a { | ||
| + | text-decoration: none; | ||
| + | font-size: 14px; | ||
| + | font-weight: bold; | ||
| + | color: #FFFFFF; | ||
| + | } | ||
| + | |||
| + | body { | ||
| + | background: white; | ||
| + | color: black; | ||
| + | margin: 0; | ||
| + | border: 0; | ||
| + | padding: 0; | ||
| + | background: url("http://www.wallpapersforvista.com/images/wallpapers/BlueKetamine1186424340-1600x1200.jpg"); | ||
| + | } | ||
| + | </style> | ||
| + | |||
| + | <body> | ||
| + | <div id="banner"><img src="http://i.imgur.com/kBtYW.png" alt="" /></div> | ||
| + | |||
| + | |||
| + | <div id="menu"> | ||
| + | <ul> | ||
| + | <li class="first"><a href="https://2012.igem.org/Team:Duke" accesskey="1" | ||
| + | title="">Home</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Duke/Team" accesskey="2" | ||
| + | title="">Meet the Team</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Duke/Project" accesskey="3" | ||
| + | title="">Project Description</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Duke/Safety" accesskey="4" | ||
| + | title="">Safety</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Duke/Notebook" accesskey="5" | ||
| + | title="">Lab Notebook</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Duke/Sponsors" accesskey="5" | ||
| + | title="">Sponsors</a></li> | ||
| + | <li><a href="https://2012.igem.org/Team:Duke/Human Practices" accesskey="6" | ||
| + | title="">Human Practices</a></li> | ||
| + | </ul> | ||
| + | |||
</div> | </div> | ||
| + | |||
| + | <!-- start page --> | ||
| + | <div id="page"> | ||
| + | <!-- start content --> | ||
| + | |||
| + | <h2 class="title"><a href="#">Engineering Optogenetic Systems in Yeast</a></h2> | ||
| + | <!-- end content --> | ||
| + | <!-- start sidebar --> | ||
| + | <div id="sidebar"> | ||
| + | <ul> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <li> | ||
| + | <h2>Useful Links</h2> | ||
| + | <ul> | ||
| + | <li><a href="http://www.genome.duke.edu/">Duke University IGSP</a></li> | ||
| + | <li><a href="http://www.bowmanbrockman.org/ncssm/">Bowman Brockman</a></li> | ||
| + | <li><a href="http://www.geneoracle.com/">Gene Oracle</a></li> | ||
| + | <li><a href="http://www.synberc.org/">SynBERC</a></li> | ||
| + | <li><a href="http://pubs.acs.org/journal/asbcd6">ACS Synethetic Biology</a></li> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | |||
| + | |||
| + | </div> | ||
</div> | </div> | ||
| + | <!-- end page --> | ||
| + | |||
| + | </body> | ||
</html> | </html> | ||
| - | |||
| - | + | == '''Overall project''' == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | Medical genetic therapy has shown promise for improved accuracy in personalized genetic therapy of conditions such as Alzheimer’s disease and cancer. However, the speed of current medical genetic screening methods is limited by time-consuming rates of cell growth and gene expression. The goal of this work, therefore, was to develop a comprehensive platform for other researchers to use in further medical genetic studies. | ||
| - | |||
| - | + | In yeast, an orthologous model of human gene function, we developed a system of two dimerizing fusion proteins to control two-hybrid mediated transcriptional activation in response to a 450 nm blue light (optogenetic) stimulus. After extensive characterization and optimization of our system, we compiled our methodologies into a physical toolkit, which contains custom yeast strains frozen in glycerol stocks, standardized plasmids, a stochastic network model, the design of a light pulse generator to induce gene expression, and a custom software package for rapid analysis of data. | |
| + | |||
| + | |||
| + | In the coming weeks, we will begin testing an application of our system by screening for orthologous suppressors of beta-amyloid that may be used in genetic therapy of Alzheimer’s disease. Our comprehensive toolkit streamlines identification of genetic therapeutic targets, and will speed progress toward personalized therapy of a variety of diseases. | ||
| + | |||
| + | |||
| + | |||
| + | The 2012 Duke iGEM Team has submitted the following parts to the Biobrick registry: | ||
| + | |||
| + | |||
| + | <groupparts>iGEM012 Duke</groupparts> | ||
| + | |||
| + | == Summary of Theory/Introduction== | ||
| + | |||
| + | Medical genetics, a field that focuses on gene therapy of medical conditions, has shown promise for identification of gene therapy targets of disease [1]. Advances in medical genetics may lead to improved personalized therapy of conditions as varied as Alzheimer’s disease, diabetes, and cancer [2–4]. However, two significant limitations exist in medical genetics that have delayed research – high cost and low speed in cell culture and gene expression [1]. | ||
| + | |||
| + | |||
| + | Immortalized human cell lines, which are typically used in medical genetics, are very accurate in determination of gene and protein function; however, gene analysis in human cells is expensive and time consuming. The fifteen human cell lines available through the Invitrogen® corporation average a cost of $1253.30; and the Invitrogen Corporation FreeStyle™ 293 Expression System, a kit that is used for analysis of protein expression in human cells, retails for $893.45 and involves a three-week waiting period before analysis. | ||
| + | |||
| + | |||
| + | High throughput screening (HTS) techniques have been applied to reduce expenditures and hasten procedures in genetics by increasing the number of samples per assay; however, the per-sample speed is still limited by the responsiveness of cell culture to researcher-imposed changes [5]. Methods to improve the accuracy and frugality of gene expression are necessary to hasten discovery in genetic screening for a variety of gene therapy targets. The purpose of our research, therefore, was to develop an improved platform for more rapid screening of gene therapeutic targets by integrating an emerging topic known as optogenetics with fast-paced flow-cytometric analysis of yeast. | ||
| + | |||
| + | |||
| + | Optogenetics, which refers to light inducible control of genes and proteins, was originally used for the regulation of synapsing by control of light-gated ion channels in neurons [6]. The inaugural works by Voigt, et al, introduced the optogenetics to cellular biology through the use of phytochromes and cryptochromes – light controllable proteins from Arabidopsis thaliana [7]. Optogenetics has shown promise for rapid, economical activation of gene expression [8]. | ||
| + | For use in fast-paced medical genetics, we chose to pair optogenetic gene activation with experimentation in yeast, a fungal microorganism. The yeast species Saccharomyces cerevisiae, is often used as a model system of eukaryotic gene function because of its homology to the human genome. The use of optogenetics in yeast offered threefold advantages of reduced cost, shortened culture time, and quantification of genetic response [9]. | ||
| + | |||
| + | |||
| + | In optogenetics, verification of gene upregulation is mediated by light output from fluorescent proteins called reporters [10]. A typical technique to detect this fluorescent output is fluorescence microscopy, however, we elected to use flow cytometric analysis, a newer technique which measures the levels of fluorescence from individual yeast cells for richer quantitative analysis of genetic feedback. Whereas fluorescence microscopy renders a single data point per yeast colony, flow cytometry yields one data point per yeast cell and typically more than 100,000 data points per colony, allowing for greater precision in gene analysis [11]. | ||
| + | For standard flow cytometric analysis, reporter feedback must be mediated by a robust promoter to ensure high levels of fluorescence. However robust gene activation in eukaryotes has been limited by long time periods of gene activation and lack of heritable genetic parts [12]. The standard of excellence for transcriptional activation in yeast cell biology is the GAL1 promoter, which mediates a one thousand fold induction of gene expression, but requires a two day waiting period for uptake of galactose sugar into the metabolic pathways of yeast. GAL1-controlled | ||
| + | genes are often introduced as plasmids, which means that the DNA can be lost after many generations [13]. We therefore integrated our optogenetic system, which is composed of a three-plasmid topology, into the chromosome of yeast using homologous recombination, ensuring a high level of gene conservation from the parent to filial generation [14]. | ||
| + | |||
| + | |||
| + | The design of our system (shown in Figure 1) is based on a yeast-two-hybrid, in which the activation domain and binding domain of the GAL4 transcription factor are split [5]. We fused GAL4(BD) to the plant cryptochrome Cry2 and VP16(AD)* [15] to the calcium binding protein CIB1. In the presence of a 450 nm wavelength blue light (optogenetic stimulus), the Cry2 and CIB1 proteins form a dimer [16,17], which unites the activation and binding domains, and activates transcription within the Gal4 upstream activation sequence. Removal of the blue light input reverses the dimerization and downregulates protein expression levels. | ||
| + | |||
| + | |||
| + | '''Figure 1. “The functionality of our optogenetic two hybrid system”.''' | ||
| + | |||
| + | [[File:Figure1Duke2012.png]] | ||
| + | |||
| + | '''Source: Adapted from unpublished work by Tucker, et al.''' | ||
| + | Dimer formation and transcription are activated with the input of a blue light (optogenetic stimulus). | ||
| + | The goal of this work was to design, characterize, and optimize an optogenetic toolkit that could serve as a platform for identification of genetic therapeutic targets for a variety of medical conditions. We utilized experimental, computational, and engineering methods to ensure comprehensiveness in development of our toolkit. | ||
| + | |||
| + | |||
| + | Our first aim was to validate the functionality of our optogenetic two-hybrid system in vivo. Our experimental objectives in this project were threefold: first, to optimize a set of reporter genes, and determine which reporter produced the most reproducible and high levels of fluorescence for feedback; second, to characterize levels of gene expression of our optogenetic system as compared to the GAL1 promoter; and third, to upload our optogenetic plasmids into the web-based BioBrick repository for further researchers to access and build upon. | ||
| + | |||
| + | |||
| + | To complement our in vivo characterization with optimization in silico, we sought to use our experimental data to develop a model of our system. Our computational objectives were: first, to develop a Gillespie stochastic system of coupled differential equations to model our network; and second, to import this data-driven model into the software TinkerCell to optimize values for variables such as light exposure in experimental use of the toolkit. | ||
| + | |||
| + | |||
| + | Our final goal was to develop our system into a practical tool for other researchers to use. We had two engineering goals in this project: first, to design a portable box to deliver specific blue light pulses to a high-throughput microarray plate; and second, to develop a software package to automate screening of flow cytometric data from our optogenetic system. | ||
| + | |||
| + | == '''Experimental Methods and Results''' == | ||
| + | |||
| + | |||
| + | During our experimentation, we constructed all plasmids within the lab through ligation of component parts and verified all plasmids through restriction enzyme digestion and PCR sequencing. All transformations into yeast used the lithium acetate/single stranded carrier DNA/polyethelene glycol method [18]. | ||
| + | |||
| + | |||
| + | '''Optimization of reporters for feedback:''' | ||
| + | |||
| + | Our first experimental goal in this work was to ensure accurate flow cytometric analysis of samples by selecting from four reporters – yEVenus (yellow), tagBFP (blue), yEGFP (green), | ||
| + | and mCherry (red) – for reproducibility and high levels of fluorescence. Reporter plasmids were transformed into yeast and level of fluorescence from the reporters was evaluated for both control samples (grown in raffinose) and induced samples (grown in galactose) by flow cytometric characterization (summarized in Figure 2). | ||
| + | |||
| + | |||
| + | '''Figure 2: “Characterization of reporter fluorescence”''' | ||
| + | |||
| + | [[File:Figure2aDuke2012.png]] | ||
| + | |||
| + | A: yEGFP (Green Fluorescent Protein) – In the inactive state, there is a high level of precision (overlap) between samples and, on average, less than 1 photon is released per yeast cell. In the active state, there is little precision, however, with levels of fluorescence broadly ranging from 50 photons/cell to 500 photons/cell. | ||
| + | OFF (Raffinose) ON (Galatose) | ||
| + | |||
| + | |||
| + | |||
| + | [[File:Figure2bDuke2012.png]] | ||
| + | |||
| + | B: tag-BFP (Blue Fluorescent Protein) – In the inactive state, peaks overlap with relatively good precision near 1 photon/cell. In the active state, there is little bimodality but there is significant spread. Fluorescence ranges from 50 to 300 photons/cell. | ||
| + | |||
| + | |||
| + | |||
| + | [[File:Figure2cDuke2012.png]] | ||
| + | |||
| + | C: mCherry (Red Fluorescent Protein) – In the inactive state, there is a high level of precision (overlap) between samples and, on average, 1 photon is released per yeast cell. In the active state, there is a high level of precision, with peak overlap and fluorescence between 400 and 600 photons per cell. | ||
| + | OFF (Raffinose) ON (Galatose) | ||
| + | OFF (Raffinose) ON (Galatose) | ||
| + | |||
| + | |||
| + | |||
| + | yEVenus (yellow fluorescent protein) was excluded from our analysis because it produced unreliable levels of fluorescence. Through analysis of characterization results, we found that the mCherry reporter was able to produce the most reliable and robust induction of fluorescence, as induction increased output from about 1 photon per cell to 500 photons per cell. By re-plating reporter patches that produced high levels of fluorescence, we selected for mCherry strains that will produce optimal fluorescent feedback in our system and in the rest of our experimentation. | ||
| + | |||
| + | Integration of optogenetic network topology at chromosomal loci | ||
| + | After characterizing reporters for use in our system, we constructed our optogenetic strains. In order to minimize loss of DNA during cell replication, we used homologous recombination to heritably insert our three-plasmid network topology into the yeast chromosome using homologous recombination. The optogenetic plasmids were first ligated into chromosomal integrating plasmids designed by Knop, et al, in which auxotrophic (amino-acid encoding) genes are split into their component 5 prime and 3 prime ends [19]. The three components of our system were transformed as three independent plasmids, and through homologous recombination GAL4(BD)-Cry2 was integrated into the HIS locus, VP16(AD)-CIB1 was integrated into the URA locus, and GAL1pr-mCherry was integrated into the TRP locus. The PEG/LiAc/Carrier DNA method that we used for our experimentation is compiled into the YeastMaker kit produced by Clontech®, which retails for $317.00. As compared to the $2146.75 in expenditures for mammalian cell lines and kits, our use of yeast reflects an 85.2% decrease in experimental cost. | ||
| + | |||
| + | '''Characterization of optogenetic system:''' | ||
| + | |||
| + | After integrating our plasmids into the chromosome of our yeast, we used the constructed strains (summarized in Table 1) to validate the functionality of our optogenetic system. Our methods for characterization of our optogenetic system were twofold: first to select for our strains using antibiotic resistance markers, and second, to run flow cytometric analysis on our samples to characterize levels of gene expression. We designed an experiment with seven control samples to minimize the effect of confounding variables on network functionality. | ||
| + | |||
| + | |||
| + | [[File:fullchartDuke2012.png]] | ||
| + | |||
| + | |||
| + | Table 1: “Summary of yeast samples constructed for antibiotic characterization of our optogenetic system” | ||
| + | The growth of the controls (without DNA) indicates the functionality of our template strains. As expected, there was no growth in the cultures with single plasmids – we found no growth unless both the Cry2 and CIB1 plasmids were transformed, indicating the accurate insertion of our CIB1 and Cry2 strains. Our MMY116 background and PJ694A experimental strains presented successful growth through resistance to HygromycinB and G418. | ||
| + | The results from our antibiotic selection assay verified that our recombinant DNA was able to confer antibiotic resistance, and therefore, our plasmids had been successfully integrated. | ||
| + | |||
| + | |||
| + | '''Standardization of plasmids in BioBrick format:''' | ||
| + | |||
| + | With the successful characterization of our optogenetic system, we expect that other researchers will seek to implement our system for a variety of applications. We developed our plasmids into interoperable parts so that other researchers could utilize our system as parts in a custom genetic network. We chose to format our plasmids as BioBricks: well-characterized, standard, biological parts which are designed to have plug-and-play capabilities for construction of biological systems [20]. The plasmids we designed will allow for rapid insertion of the user’s gene of interest. The design and characterization data for our BioBrick plasmids were uploaded to the web-based Repository of Standard Biological Parts for other researchers to access, and the list of parts are summarized in Table 2. | ||
| + | Table 2. “A listing of BioBricks published to the Registry of Standard Biological Parts.” | ||
| + | We uploaded all of the plasmids that we used in our project, which had not already been characterized, to the web-based Registry of Standard Biological Parts. Researchers will be able to easily access our parts and integrate these components into their own biological systems. | ||
| + | |||
| + | |||
| + | [[File:BiobricklistDuke2012.png]] | ||
| + | |||
| + | =='''Computational Modeling & Results'''== | ||
| + | Stochastic modeling of optogenetic network: | ||
| + | Although we were able to complete extensive development and characterization of our system in vivo, we were unable to optimize the performance of our system experimentally. We therefore chose to construct a stochastic model of our system to predict network behavior, and optimize the performance of the system for changes in certain variables. To ensure accuracy in our model, we used the experimental data we gathered and information from the Registry of Standard Biological Parts. The data gathered included information about promoter strength, transcription factor affinity, and protein degradation rate, and were used to develop a stochastic model composed of a system of coupled differential equations. However, in our system, we developed two biological parts that had not been characterized – the interacting CIB1 and Cry2 proteins. Rather than using an arbitrary default value, we developed a Gillespie simulation, a statistically correct trajectory of a stochastic equation, to characterize the dimerizing efficiency of these two proteins. In our model, we inserted the coupled differential equations described by Gillespie, et al, that are used to describe the kinematics of dimerizing chemical reactions of the type A+B → AB → A+B [21]. | ||
| + | We imported these data into TinkerCell, a computer aided design software tool, which enabled plug-and-play construction of our network topology [22]. Using TinkerCell, we designed a graphical representation of our network (shown in Figure 4) by taking the network topology from plasmid maps we designed in Sequence Builder. The model takes into account several input variables, such as amount of blue light (representing length of time), degradation rate, and probability of dimer formation. | ||
| + | |||
| + | |||
| + | [[File:Figure4Duke2012.png]] | ||
| + | |||
| + | |||
| + | Figure 4: “Network topology of the optogenetic two-hybrid system” - Our genetic network topology consists of a three plasmid design. (A) The two constructs on the left (top and bottom) represent the two coding sequences for Cry2 and CIB1, the dimerizing proteins of the two-hybrid system. In the presence of blue light these two proteins form a dimer as seen in (B). This dimer induces the GAL1 Promoter, activating transcription of the upstream activation sequence (C). This upregulates our protein of interest and mCherry reporter fusion protein, allowing for flow cytometric analysis of protein expression. | ||
| + | |||
| + | |||
| + | [[File:Figure5Duke2012.png]] | ||
| + | |||
| + | |||
| + | Figure 5: “Computational characterization of network behavior” - The solution to our system of coupled differential equations yielded a stochastic model of the network behavior. As expected, the system demonstrated the ability to alternate between to states with the input of blue light stimulus. CIB 1 and Cry2 are weakly upregulated without blue light (Graph A), whereas with blue light, the protein of interest and mCherry fusion is strongly upregulated. Dimer formation and depletion of endogenous Cry2 and CIB1 in the presence of blue light is consistent with the dimer-driven mechanism of our optogenetic system. | ||
| + | |||
| + | |||
| + | First, we constructed a deterministic model to determine the natural steady state without light inducers of gene expression. Without any inducers, the dimerizing proteins VP16(AD)-CIB1 and GAL4(BD)-Cry2 are weakly upregulated, whereas the protein of interest, the reporter, and the dimer are maintained at a basal state (Figure 5: Graph A). In our model, application of blue light caused dimer formation, depleting free-floating CIB1 and Cry2 and activating GAL4-mediated transcription of the reporter and gene of interest fusion (Figure 5: Graph B). The light-mediated upregulation of our protein of interest, the mCherry reporter, and the dimer formation is visible in Graph B. | ||
| + | |||
| + | The strong responsiveness to blue light and the low levels of activation in the basal state indicated that our computational model exhibits the behavior that we observed in vivo. We used this stochastic model for experimentation in silico – in particular, we conducted experimentation that required extremely high number of samples to determine the seconds of blue light that optimizes levels of protein expression. In our model, 20.0 - 22.0 seconds of blue light exposure optimized the maximum (induced) state among simulations. This time period for gene activation represents a 6500 fold increase in the speed of gene activation as compared to the 36 hour period of galactose exposure for the GAL1 promoter [13]. | ||
| + | The characteristics of the network – rapid, extensive gene induction – matched results from our in vivo characterization. This model is therefore a useful aspect of our final toolkit that researchers can use for pre-experimental hypothesis. By doing a preliminary screen in silico, our computational model can save researchers time in analyzing large sample sets. | ||
| + | |||
| + | == '''Software Engineering Methods & Results''' == | ||
| + | |||
| + | '''Automation of flow cytometric data screening:''' | ||
| + | Throughout much of our experimental work to characterize our optogenetic system, we found that the data gathered from the flow cytometer took nearly a full day to analyze completely. Although our system allows for more rapid experimentation in medical genetics, the speed of discovery is limited by how quickly the flow cytometric data can be analyzed. We found that the typical procedure for analysis of data involved moving files, developing graphs, analyzing graphs, and looking up human orthologs for promising yeast genes over a time period of several hours. | ||
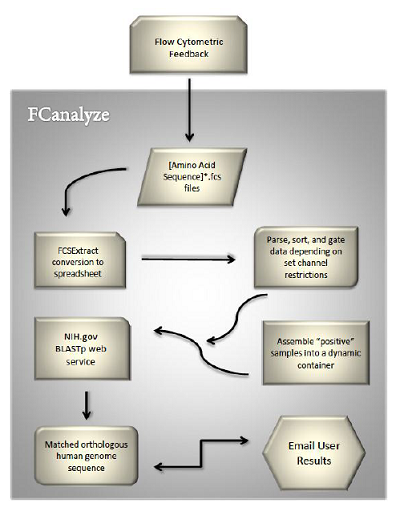
| + | In order to streamline the genetic screening process from start to finish, we developed a computer program from scratch to complement our experimental platform. The user saves their .fcs (flow cytometry standard output) files into a single folder and inputs the directory in which the files are located. We utilized FCSExtract, an opensource software, to convert .fcs (flow cytometer) files into .csv (spreadsheet) files. Our C++ software then uses dynamic memory to store and sort over 1.02e6 bytes (about 1 MB) of data per .csv file. The final output from our program is an email listing of the human orthologs of the yeast genes that were positives in the screening process. The functionality of our program is summarized in Figure 2. | ||
| + | |||
| + | [[File:FCAnalyzeDiagram.png]] | ||
| + | |||
| + | We verified the functionality of our software using the flow cytometric data we gathered from our characterization of the optogenetic system. We selected 50 files for analysis. Our software successfully gated the files and aligned them into a separate container in, on average, 1.15 seconds. The orthologus Amino Acid sequence corresponding to each sample with gene activation was returned from the NIH BLASTp server in on average, 2.3 seconds. In our experimental work, analysis of flow cytometric data from 50 files required 7.5 hours of work. Our software was able to complete the task in 3.45 seconds, which represents a 7800-fold increase in the speed of flow cytometric data analysis [22]. | ||
| + | |||
| + | == '''Future Work''' == | ||
| + | |||
| + | |||
| + | So far, we have designed transformation plasmids in SeqBuilder so the end user of our toolkit could easily integrate a gene-of-interest into our custom yeast strains in two steps using a custom transformation plasmid. A user’s gene-of-interest will be integrated into the TRP chromosomal locus by homologous recombination, and flanked by a reporter with a glycine linkage helix. The translated protein will be the users protein-of-interest with a reporter “tag,” allowing for flow cytometric analysis of levels of gene expression [11]. We have submitted a funding proposal to order these designed plasmids de novo. | ||
| + | We proposed the design of one potential application of our toolkit for loss of function screening. In a typical LOF assay, a users substance of interest is introduced into the yeast gene deletion set, a set of 6000 different gene knockdown yeast strains [24]. This tool is traditionally used to identify genetic suppressors of a chemical compound – for example, identifying cancer related genes that confer resistance to chemotherapeutic drugs such as Paclitaxel [2,3]. | ||
| + | |||
| + | In the coming weeks, we plan to acquire a yeast gene deletion set through personal correspondence and transform it with our optogenetic plasmids and a GAL1 controlled gene-of-interest mCherry fusion. The transformation plasmid we have designed will integrate a users gene of interest into our custom deletion set. Our variation of the traditional assay will allow for rapid screening for repressors of not only chemical compounds, but also a user’s fluorescence tagged gene of interest. The samples will be screened in a flow cytometer, and the samples for which levels of fluorescence (and therefore, the gene of interest) are extremely high indicate that the deleted gene is a repressor of a user’s gene of interest. Conversely, we can screen for activators of a gene of interest by evaluating deletions that cause extremely low levels of expression of the gene-of-interest. The process of data analysis from our LOF assay is entirely automated by our software package, which can take raw flow cytometry data and rapidly identify human orthologs of the corresponding yeast suppressor or activator. The optimization of LOF protocols proposed by Giorgini, et al, by using our optogenetic tools may hold a number of other applications for preliminary identification of genes that may be used in gene therapy [25]. | ||
| + | |||
| + | One of the most significant applications of this technique would be for identification of genetic and enzymatic suppressors of Beta-Amyloid, a causal protein in the onset of Alzheimer’s disease. Although Beta-Amyloid forms a plaque that ultimately leads to neurodegeneration, it is also necessary in small amounts for normal brain function. For improved treatment of Alzheimer’s disease, a weak suppressor needs to be identified that holds Beta-Amyloid at a median level. For Alzheimer’s patients, genetic therapy using weak enzymatic suppressors of Beta-Amyloid has the potential to halt and regress disease progression [4]. | ||
| + | |||
| + | Unlike traditional loss of function assays, which can only identify if a gene is a suppressor or not, using our more precise, dosage dependent optogenetic methods, we can characterize suppressor dynamics. The use of our optogenetic system can allow us to identify weak suppressors of beta-amyloid by screening for suppressors that yield median levels of beta-amyloid fluorescence. We have already identified the yeast ortholog of Beta-Amyloid by using the NIH BLASTp web service. We can conceivably begin screening for suppressors of Beta-Amyloid in the coming weeks when we acquire our transformation plasmid and a yeast gene deletion set. The platform we have developed in this work is not limited to Alzheimer’s disease, however; and will be used for identification of gene therapy targets for as varied conditions as Huntington’s disease, cancer, and diabetes [2,3]. | ||
| + | |||
| + | =='''Conclusion''' == | ||
| + | |||
| + | In this work, we described the development, characterization, and optimization of an optogenetic toolkit for use in medical genetic studies. We were able to make improvements in the speed, cost, and accuracy of genetic screening by integrating yeast flow cytometric analysis with optogenetic control of transcription. We optimized and selected for three different fluorescent proteins that produced reliable levels of feedback for use in our toolkit and characterized our optogenetic system. We found that our optogenetic system produced equally reliable levels of gene induction as the widely-used GAL1 promoter, with a 22.0 second response time. To complement our experimental utlities, we utilized the modeling software TinkerCell to optimize values for numerous experimental variables in our system. Finally, we developed a C++ software program to take flow cytometry output from our system and process the data, streamlining the process of data analysis that limits current high-throughput techniques. Our toolkit was able to make a 5900-fold improvement in the speed of gene activation in medical genetic assays and a 7800-fold increase in the speed of analysis of flow cytometric data. Our experimental methods also yielded an 85.2% decrease in expenditures as compared to the use of mammalian cell lines. In the coming weeks, we will develop a custom yeast gene deletion set and loss of function assay as a proof-of-concept of our system, and begin testing an application of our toolkit by screening for orthologous suppressors of beta-amyloid that may be used in genetic therapy of Alzheimer’s disease. The construction of our toolkit will prompt advances in gene therapy and is a major step towards personalized therapy of a variety of diseases. | ||
| + | |||
| + | == '''References''' == | ||
| + | |||
| + | [1] S. Goverdhana, M. Puntel, W. Xiong, J. M. Zirger, C. Barcia, J. F. Curtin, E. B. Soffer, S. Mondkar, G. D. King, J. Hu, S. a Sciascia, M. Candolfi, D. S. Greengold, P. R. Lowenstein, and M. G. Castro, Molecular Therapy : the Journal of the American Society of Gene Therapy 12, 189 (2005). | ||
| + | |||
| + | [2] V. N. Ngo, R. E. Davis, L. Lamy, X. Yu, H. Zhao, G. Lenz, L. T. Lam, S. Dave, L. Yang, J. Powell, and L. M. Staudt, Nature 441, 106 (2006). | ||
| + | |||
| + | [3] W. Farris, S. Mansourian, M. a. Leissring, E. a. Eckman, L. Bertram, C. B. Eckman, R. E. Tanzi, and D. J. Selkoe, The American Journal of Pathology 164, 1425 (2004). | ||
| + | |||
| + | [4] J. J. Palop and L. Mucke, Nature Neuroscience 13, 812 (2010). | ||
| + | |||
| + | [5] J. Chen, M. B. Carter, B. S. Edwards, H. Cai, and L. a Sklar, Cytometry. Part A : the Journal of the International Society for Analytical Cytology 81, 90 (2012). | ||
| + | [6] L. Fenno, O. Yizhar, and K. Deisseroth, Annual Review of Neuroscience 34, 389 (2011). | ||
| + | [7] J. J. Tabor, A. Levskaya, and C. a Voigt, Journal of Molecular Biology 405, 315 (2011). | ||
| + | [8] L. Gardner and A. Deiters, Current Opinion in Chemical Biology 16, 292 (2012). | ||
| + | [9] F. Sherman, Methods Enzymol 350, (1991). | ||
| + | [10] J. Toettcher, C. Voigt, O. Weiner, and W. Lim, Nature Methods 8, 35 (2010). | ||
| + | [11] C. Mateus and S. V. Avery, Yeast (Chichester, England) 16, 1313 (2000). | ||
| - | + | [12] M. Manning and L. Hudgins, Genetics in Medicine : Official Journal of the American College of Medical Genetics 12, 742 (2010). | |
| + | [13] D. Lohr, P. Venkov, and J. Zlatanova, The FASEB Journal 777 (1995). | ||
| + | [14] H. Ma, S. Kunes, P. Schatz, and D. Botstein, Gene 201 (1987). | ||
| + | [15] D. B. Hall and K. Struhl, The Journal of Biological Chemistry 277, 46043 (2002). | ||
| + | [16] H. Liu, X. Yu, K. Li, and J. Klejnot, Science … 322, 1535 (2008). | ||
| - | + | [17] C. Viappiani, S. Nonell, and M. Andreas, (2010). | |
| + | [18] R. D. Gietz and R. a Woods, Methods in Enzymology 350, 87 (2002). | ||
| + | Page 20 | ||
| + | [19] C. Janke, M. M. Magiera, N. Rathfelder, C. Taxis, S. Reber, H. Maekawa, A. Moreno-Borchart, G. Doenges, E. Schwob, E. Schiebel, and M. Knop, Yeast (Chichester, England) 21, 947 (2004). | ||
| + | [20] C. D. Smolke, Nature Biotechnology 27, 1099 (2009). | ||
| + | [21] D. T. Gillespie, Annual Review of Physical Chemistry 58, 35 (2007). | ||
| - | + | [22] D. Chandran, F. T. Bergmann, and H. M. Sauro, Journal of Biological Engineering 3, 19 (2009). | |
| + | [23] M. a Naivar, M. E. Wilder, R. C. Habbersett, T. a Woods, D. S. Sebba, J. P. Nolan, and S. W. Graves, Cytometry. Part A : the Journal of the International Society for Analytical Cytology 75, 979 (2009). | ||
| + | [24] C. B. Brachmann, a Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke, Yeast (Chichester, England) 14, 115 (1998). | ||
| + | [25] F. Giorgini, P. Guidetti, Q. Nguyen, S. C. Bennett, and P. J. Muchowski, Nature Genetics 37, 526 (2005). | ||
| - | + | [26] E. J. Olson and J. J. Tabor, Current Opinion in Chemical Biology 16, 300 (2012). | |
| + | [27] R. M. Johnson, C. Allen, S. D. Melman, A. Waller, S. M. Young, L. a Sklar, and K. J. Parra, Analytical Biochemistry 398, 203 (2010). | ||
| + | [28] G. Charvin, F. R. Cross, and E. D. Siggia, PloS One 3, e1468 (2008). | ||
| + | [29] N. E. Buchler and F. R. Cross, Molecular Systems Biology 5, 272 (2009). | ||
| - | + | [30] S. Shimizu-Sato, E. Huq, J. M. Tepperman, and P. H. Quail, Nature Biotechnology 20, 1041 (2002). | |
Latest revision as of 21:17, 15 October 2012
Contents |
Overall project
Medical genetic therapy has shown promise for improved accuracy in personalized genetic therapy of conditions such as Alzheimer’s disease and cancer. However, the speed of current medical genetic screening methods is limited by time-consuming rates of cell growth and gene expression. The goal of this work, therefore, was to develop a comprehensive platform for other researchers to use in further medical genetic studies.
In yeast, an orthologous model of human gene function, we developed a system of two dimerizing fusion proteins to control two-hybrid mediated transcriptional activation in response to a 450 nm blue light (optogenetic) stimulus. After extensive characterization and optimization of our system, we compiled our methodologies into a physical toolkit, which contains custom yeast strains frozen in glycerol stocks, standardized plasmids, a stochastic network model, the design of a light pulse generator to induce gene expression, and a custom software package for rapid analysis of data.
In the coming weeks, we will begin testing an application of our system by screening for orthologous suppressors of beta-amyloid that may be used in genetic therapy of Alzheimer’s disease. Our comprehensive toolkit streamlines identification of genetic therapeutic targets, and will speed progress toward personalized therapy of a variety of diseases.
The 2012 Duke iGEM Team has submitted the following parts to the Biobrick registry:
<groupparts>iGEM012 Duke</groupparts>
Summary of Theory/Introduction
Medical genetics, a field that focuses on gene therapy of medical conditions, has shown promise for identification of gene therapy targets of disease [1]. Advances in medical genetics may lead to improved personalized therapy of conditions as varied as Alzheimer’s disease, diabetes, and cancer [2–4]. However, two significant limitations exist in medical genetics that have delayed research – high cost and low speed in cell culture and gene expression [1].
Immortalized human cell lines, which are typically used in medical genetics, are very accurate in determination of gene and protein function; however, gene analysis in human cells is expensive and time consuming. The fifteen human cell lines available through the Invitrogen® corporation average a cost of $1253.30; and the Invitrogen Corporation FreeStyle™ 293 Expression System, a kit that is used for analysis of protein expression in human cells, retails for $893.45 and involves a three-week waiting period before analysis.
High throughput screening (HTS) techniques have been applied to reduce expenditures and hasten procedures in genetics by increasing the number of samples per assay; however, the per-sample speed is still limited by the responsiveness of cell culture to researcher-imposed changes [5]. Methods to improve the accuracy and frugality of gene expression are necessary to hasten discovery in genetic screening for a variety of gene therapy targets. The purpose of our research, therefore, was to develop an improved platform for more rapid screening of gene therapeutic targets by integrating an emerging topic known as optogenetics with fast-paced flow-cytometric analysis of yeast.
Optogenetics, which refers to light inducible control of genes and proteins, was originally used for the regulation of synapsing by control of light-gated ion channels in neurons [6]. The inaugural works by Voigt, et al, introduced the optogenetics to cellular biology through the use of phytochromes and cryptochromes – light controllable proteins from Arabidopsis thaliana [7]. Optogenetics has shown promise for rapid, economical activation of gene expression [8].
For use in fast-paced medical genetics, we chose to pair optogenetic gene activation with experimentation in yeast, a fungal microorganism. The yeast species Saccharomyces cerevisiae, is often used as a model system of eukaryotic gene function because of its homology to the human genome. The use of optogenetics in yeast offered threefold advantages of reduced cost, shortened culture time, and quantification of genetic response [9].
In optogenetics, verification of gene upregulation is mediated by light output from fluorescent proteins called reporters [10]. A typical technique to detect this fluorescent output is fluorescence microscopy, however, we elected to use flow cytometric analysis, a newer technique which measures the levels of fluorescence from individual yeast cells for richer quantitative analysis of genetic feedback. Whereas fluorescence microscopy renders a single data point per yeast colony, flow cytometry yields one data point per yeast cell and typically more than 100,000 data points per colony, allowing for greater precision in gene analysis [11].
For standard flow cytometric analysis, reporter feedback must be mediated by a robust promoter to ensure high levels of fluorescence. However robust gene activation in eukaryotes has been limited by long time periods of gene activation and lack of heritable genetic parts [12]. The standard of excellence for transcriptional activation in yeast cell biology is the GAL1 promoter, which mediates a one thousand fold induction of gene expression, but requires a two day waiting period for uptake of galactose sugar into the metabolic pathways of yeast. GAL1-controlled
genes are often introduced as plasmids, which means that the DNA can be lost after many generations [13]. We therefore integrated our optogenetic system, which is composed of a three-plasmid topology, into the chromosome of yeast using homologous recombination, ensuring a high level of gene conservation from the parent to filial generation [14].
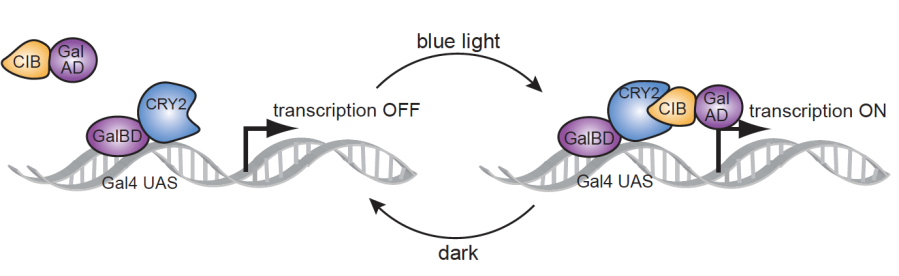
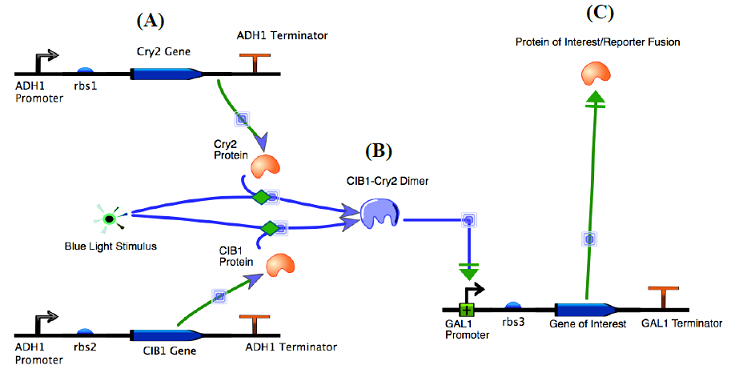
The design of our system (shown in Figure 1) is based on a yeast-two-hybrid, in which the activation domain and binding domain of the GAL4 transcription factor are split [5]. We fused GAL4(BD) to the plant cryptochrome Cry2 and VP16(AD)* [15] to the calcium binding protein CIB1. In the presence of a 450 nm wavelength blue light (optogenetic stimulus), the Cry2 and CIB1 proteins form a dimer [16,17], which unites the activation and binding domains, and activates transcription within the Gal4 upstream activation sequence. Removal of the blue light input reverses the dimerization and downregulates protein expression levels.
Figure 1. “The functionality of our optogenetic two hybrid system”.
Source: Adapted from unpublished work by Tucker, et al. Dimer formation and transcription are activated with the input of a blue light (optogenetic stimulus). The goal of this work was to design, characterize, and optimize an optogenetic toolkit that could serve as a platform for identification of genetic therapeutic targets for a variety of medical conditions. We utilized experimental, computational, and engineering methods to ensure comprehensiveness in development of our toolkit.
Our first aim was to validate the functionality of our optogenetic two-hybrid system in vivo. Our experimental objectives in this project were threefold: first, to optimize a set of reporter genes, and determine which reporter produced the most reproducible and high levels of fluorescence for feedback; second, to characterize levels of gene expression of our optogenetic system as compared to the GAL1 promoter; and third, to upload our optogenetic plasmids into the web-based BioBrick repository for further researchers to access and build upon.
To complement our in vivo characterization with optimization in silico, we sought to use our experimental data to develop a model of our system. Our computational objectives were: first, to develop a Gillespie stochastic system of coupled differential equations to model our network; and second, to import this data-driven model into the software TinkerCell to optimize values for variables such as light exposure in experimental use of the toolkit.
Our final goal was to develop our system into a practical tool for other researchers to use. We had two engineering goals in this project: first, to design a portable box to deliver specific blue light pulses to a high-throughput microarray plate; and second, to develop a software package to automate screening of flow cytometric data from our optogenetic system.
Experimental Methods and Results
During our experimentation, we constructed all plasmids within the lab through ligation of component parts and verified all plasmids through restriction enzyme digestion and PCR sequencing. All transformations into yeast used the lithium acetate/single stranded carrier DNA/polyethelene glycol method [18].
Optimization of reporters for feedback:
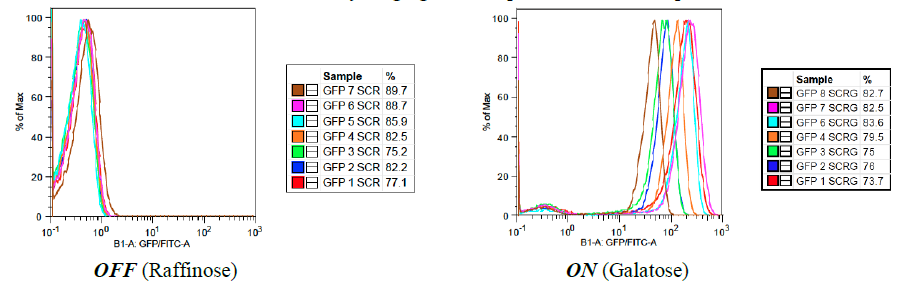
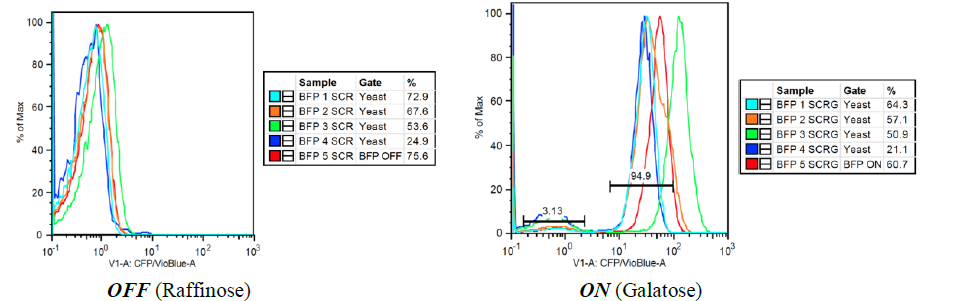
Our first experimental goal in this work was to ensure accurate flow cytometric analysis of samples by selecting from four reporters – yEVenus (yellow), tagBFP (blue), yEGFP (green), and mCherry (red) – for reproducibility and high levels of fluorescence. Reporter plasmids were transformed into yeast and level of fluorescence from the reporters was evaluated for both control samples (grown in raffinose) and induced samples (grown in galactose) by flow cytometric characterization (summarized in Figure 2).
Figure 2: “Characterization of reporter fluorescence”
A: yEGFP (Green Fluorescent Protein) – In the inactive state, there is a high level of precision (overlap) between samples and, on average, less than 1 photon is released per yeast cell. In the active state, there is little precision, however, with levels of fluorescence broadly ranging from 50 photons/cell to 500 photons/cell. OFF (Raffinose) ON (Galatose)
B: tag-BFP (Blue Fluorescent Protein) – In the inactive state, peaks overlap with relatively good precision near 1 photon/cell. In the active state, there is little bimodality but there is significant spread. Fluorescence ranges from 50 to 300 photons/cell.
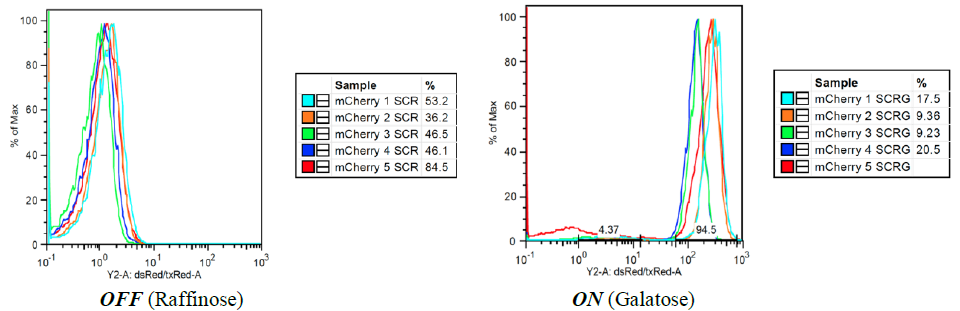
C: mCherry (Red Fluorescent Protein) – In the inactive state, there is a high level of precision (overlap) between samples and, on average, 1 photon is released per yeast cell. In the active state, there is a high level of precision, with peak overlap and fluorescence between 400 and 600 photons per cell. OFF (Raffinose) ON (Galatose) OFF (Raffinose) ON (Galatose)
yEVenus (yellow fluorescent protein) was excluded from our analysis because it produced unreliable levels of fluorescence. Through analysis of characterization results, we found that the mCherry reporter was able to produce the most reliable and robust induction of fluorescence, as induction increased output from about 1 photon per cell to 500 photons per cell. By re-plating reporter patches that produced high levels of fluorescence, we selected for mCherry strains that will produce optimal fluorescent feedback in our system and in the rest of our experimentation.
Integration of optogenetic network topology at chromosomal loci After characterizing reporters for use in our system, we constructed our optogenetic strains. In order to minimize loss of DNA during cell replication, we used homologous recombination to heritably insert our three-plasmid network topology into the yeast chromosome using homologous recombination. The optogenetic plasmids were first ligated into chromosomal integrating plasmids designed by Knop, et al, in which auxotrophic (amino-acid encoding) genes are split into their component 5 prime and 3 prime ends [19]. The three components of our system were transformed as three independent plasmids, and through homologous recombination GAL4(BD)-Cry2 was integrated into the HIS locus, VP16(AD)-CIB1 was integrated into the URA locus, and GAL1pr-mCherry was integrated into the TRP locus. The PEG/LiAc/Carrier DNA method that we used for our experimentation is compiled into the YeastMaker kit produced by Clontech®, which retails for $317.00. As compared to the $2146.75 in expenditures for mammalian cell lines and kits, our use of yeast reflects an 85.2% decrease in experimental cost.
Characterization of optogenetic system:
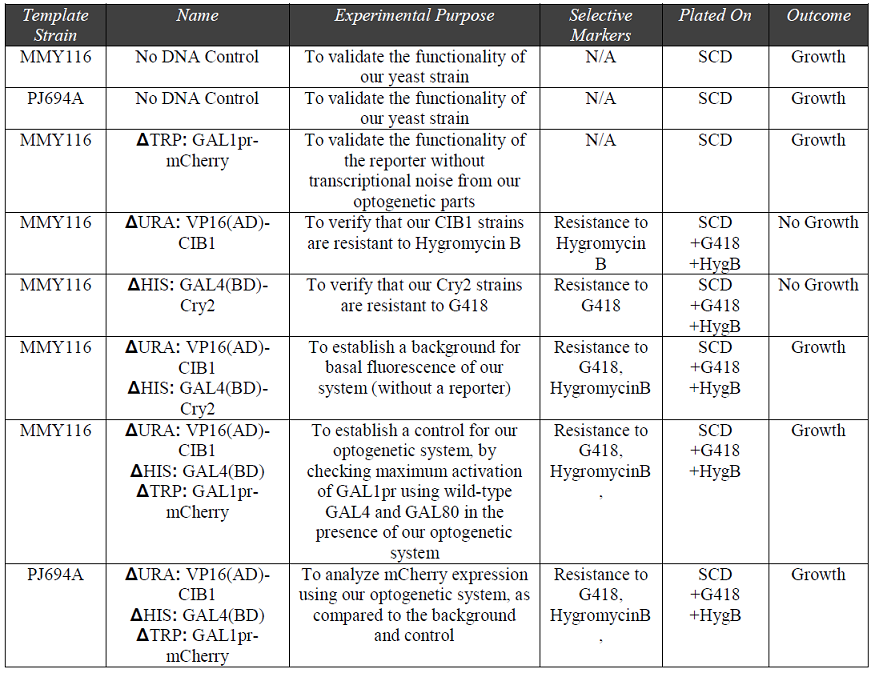
After integrating our plasmids into the chromosome of our yeast, we used the constructed strains (summarized in Table 1) to validate the functionality of our optogenetic system. Our methods for characterization of our optogenetic system were twofold: first to select for our strains using antibiotic resistance markers, and second, to run flow cytometric analysis on our samples to characterize levels of gene expression. We designed an experiment with seven control samples to minimize the effect of confounding variables on network functionality.
Table 1: “Summary of yeast samples constructed for antibiotic characterization of our optogenetic system”
The growth of the controls (without DNA) indicates the functionality of our template strains. As expected, there was no growth in the cultures with single plasmids – we found no growth unless both the Cry2 and CIB1 plasmids were transformed, indicating the accurate insertion of our CIB1 and Cry2 strains. Our MMY116 background and PJ694A experimental strains presented successful growth through resistance to HygromycinB and G418.
The results from our antibiotic selection assay verified that our recombinant DNA was able to confer antibiotic resistance, and therefore, our plasmids had been successfully integrated.
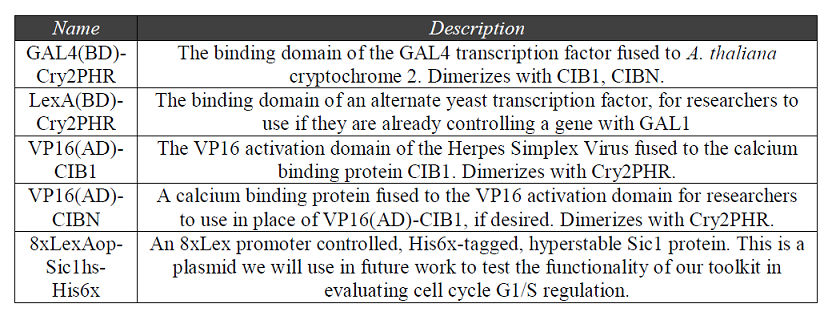
Standardization of plasmids in BioBrick format:
With the successful characterization of our optogenetic system, we expect that other researchers will seek to implement our system for a variety of applications. We developed our plasmids into interoperable parts so that other researchers could utilize our system as parts in a custom genetic network. We chose to format our plasmids as BioBricks: well-characterized, standard, biological parts which are designed to have plug-and-play capabilities for construction of biological systems [20]. The plasmids we designed will allow for rapid insertion of the user’s gene of interest. The design and characterization data for our BioBrick plasmids were uploaded to the web-based Repository of Standard Biological Parts for other researchers to access, and the list of parts are summarized in Table 2. Table 2. “A listing of BioBricks published to the Registry of Standard Biological Parts.” We uploaded all of the plasmids that we used in our project, which had not already been characterized, to the web-based Registry of Standard Biological Parts. Researchers will be able to easily access our parts and integrate these components into their own biological systems.
Computational Modeling & Results
Stochastic modeling of optogenetic network: Although we were able to complete extensive development and characterization of our system in vivo, we were unable to optimize the performance of our system experimentally. We therefore chose to construct a stochastic model of our system to predict network behavior, and optimize the performance of the system for changes in certain variables. To ensure accuracy in our model, we used the experimental data we gathered and information from the Registry of Standard Biological Parts. The data gathered included information about promoter strength, transcription factor affinity, and protein degradation rate, and were used to develop a stochastic model composed of a system of coupled differential equations. However, in our system, we developed two biological parts that had not been characterized – the interacting CIB1 and Cry2 proteins. Rather than using an arbitrary default value, we developed a Gillespie simulation, a statistically correct trajectory of a stochastic equation, to characterize the dimerizing efficiency of these two proteins. In our model, we inserted the coupled differential equations described by Gillespie, et al, that are used to describe the kinematics of dimerizing chemical reactions of the type A+B → AB → A+B [21]. We imported these data into TinkerCell, a computer aided design software tool, which enabled plug-and-play construction of our network topology [22]. Using TinkerCell, we designed a graphical representation of our network (shown in Figure 4) by taking the network topology from plasmid maps we designed in Sequence Builder. The model takes into account several input variables, such as amount of blue light (representing length of time), degradation rate, and probability of dimer formation.
Figure 4: “Network topology of the optogenetic two-hybrid system” - Our genetic network topology consists of a three plasmid design. (A) The two constructs on the left (top and bottom) represent the two coding sequences for Cry2 and CIB1, the dimerizing proteins of the two-hybrid system. In the presence of blue light these two proteins form a dimer as seen in (B). This dimer induces the GAL1 Promoter, activating transcription of the upstream activation sequence (C). This upregulates our protein of interest and mCherry reporter fusion protein, allowing for flow cytometric analysis of protein expression.
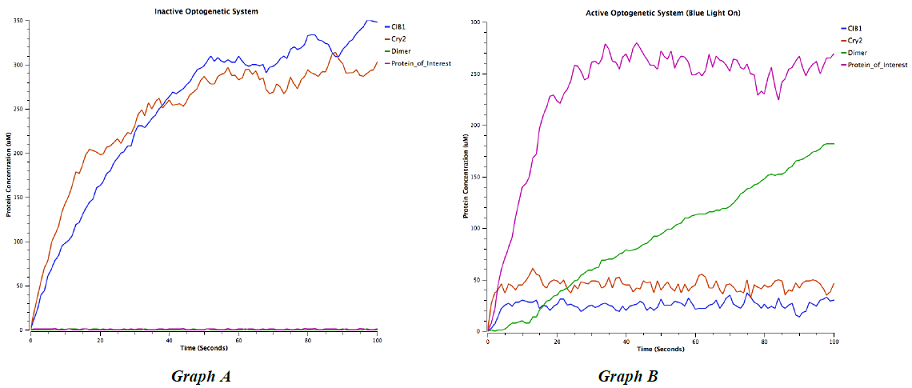
Figure 5: “Computational characterization of network behavior” - The solution to our system of coupled differential equations yielded a stochastic model of the network behavior. As expected, the system demonstrated the ability to alternate between to states with the input of blue light stimulus. CIB 1 and Cry2 are weakly upregulated without blue light (Graph A), whereas with blue light, the protein of interest and mCherry fusion is strongly upregulated. Dimer formation and depletion of endogenous Cry2 and CIB1 in the presence of blue light is consistent with the dimer-driven mechanism of our optogenetic system.
First, we constructed a deterministic model to determine the natural steady state without light inducers of gene expression. Without any inducers, the dimerizing proteins VP16(AD)-CIB1 and GAL4(BD)-Cry2 are weakly upregulated, whereas the protein of interest, the reporter, and the dimer are maintained at a basal state (Figure 5: Graph A). In our model, application of blue light caused dimer formation, depleting free-floating CIB1 and Cry2 and activating GAL4-mediated transcription of the reporter and gene of interest fusion (Figure 5: Graph B). The light-mediated upregulation of our protein of interest, the mCherry reporter, and the dimer formation is visible in Graph B.
The strong responsiveness to blue light and the low levels of activation in the basal state indicated that our computational model exhibits the behavior that we observed in vivo. We used this stochastic model for experimentation in silico – in particular, we conducted experimentation that required extremely high number of samples to determine the seconds of blue light that optimizes levels of protein expression. In our model, 20.0 - 22.0 seconds of blue light exposure optimized the maximum (induced) state among simulations. This time period for gene activation represents a 6500 fold increase in the speed of gene activation as compared to the 36 hour period of galactose exposure for the GAL1 promoter [13]. The characteristics of the network – rapid, extensive gene induction – matched results from our in vivo characterization. This model is therefore a useful aspect of our final toolkit that researchers can use for pre-experimental hypothesis. By doing a preliminary screen in silico, our computational model can save researchers time in analyzing large sample sets.
Software Engineering Methods & Results
Automation of flow cytometric data screening: Throughout much of our experimental work to characterize our optogenetic system, we found that the data gathered from the flow cytometer took nearly a full day to analyze completely. Although our system allows for more rapid experimentation in medical genetics, the speed of discovery is limited by how quickly the flow cytometric data can be analyzed. We found that the typical procedure for analysis of data involved moving files, developing graphs, analyzing graphs, and looking up human orthologs for promising yeast genes over a time period of several hours. In order to streamline the genetic screening process from start to finish, we developed a computer program from scratch to complement our experimental platform. The user saves their .fcs (flow cytometry standard output) files into a single folder and inputs the directory in which the files are located. We utilized FCSExtract, an opensource software, to convert .fcs (flow cytometer) files into .csv (spreadsheet) files. Our C++ software then uses dynamic memory to store and sort over 1.02e6 bytes (about 1 MB) of data per .csv file. The final output from our program is an email listing of the human orthologs of the yeast genes that were positives in the screening process. The functionality of our program is summarized in Figure 2.
We verified the functionality of our software using the flow cytometric data we gathered from our characterization of the optogenetic system. We selected 50 files for analysis. Our software successfully gated the files and aligned them into a separate container in, on average, 1.15 seconds. The orthologus Amino Acid sequence corresponding to each sample with gene activation was returned from the NIH BLASTp server in on average, 2.3 seconds. In our experimental work, analysis of flow cytometric data from 50 files required 7.5 hours of work. Our software was able to complete the task in 3.45 seconds, which represents a 7800-fold increase in the speed of flow cytometric data analysis [22].
Future Work
So far, we have designed transformation plasmids in SeqBuilder so the end user of our toolkit could easily integrate a gene-of-interest into our custom yeast strains in two steps using a custom transformation plasmid. A user’s gene-of-interest will be integrated into the TRP chromosomal locus by homologous recombination, and flanked by a reporter with a glycine linkage helix. The translated protein will be the users protein-of-interest with a reporter “tag,” allowing for flow cytometric analysis of levels of gene expression [11]. We have submitted a funding proposal to order these designed plasmids de novo. We proposed the design of one potential application of our toolkit for loss of function screening. In a typical LOF assay, a users substance of interest is introduced into the yeast gene deletion set, a set of 6000 different gene knockdown yeast strains [24]. This tool is traditionally used to identify genetic suppressors of a chemical compound – for example, identifying cancer related genes that confer resistance to chemotherapeutic drugs such as Paclitaxel [2,3].
In the coming weeks, we plan to acquire a yeast gene deletion set through personal correspondence and transform it with our optogenetic plasmids and a GAL1 controlled gene-of-interest mCherry fusion. The transformation plasmid we have designed will integrate a users gene of interest into our custom deletion set. Our variation of the traditional assay will allow for rapid screening for repressors of not only chemical compounds, but also a user’s fluorescence tagged gene of interest. The samples will be screened in a flow cytometer, and the samples for which levels of fluorescence (and therefore, the gene of interest) are extremely high indicate that the deleted gene is a repressor of a user’s gene of interest. Conversely, we can screen for activators of a gene of interest by evaluating deletions that cause extremely low levels of expression of the gene-of-interest. The process of data analysis from our LOF assay is entirely automated by our software package, which can take raw flow cytometry data and rapidly identify human orthologs of the corresponding yeast suppressor or activator. The optimization of LOF protocols proposed by Giorgini, et al, by using our optogenetic tools may hold a number of other applications for preliminary identification of genes that may be used in gene therapy [25].
One of the most significant applications of this technique would be for identification of genetic and enzymatic suppressors of Beta-Amyloid, a causal protein in the onset of Alzheimer’s disease. Although Beta-Amyloid forms a plaque that ultimately leads to neurodegeneration, it is also necessary in small amounts for normal brain function. For improved treatment of Alzheimer’s disease, a weak suppressor needs to be identified that holds Beta-Amyloid at a median level. For Alzheimer’s patients, genetic therapy using weak enzymatic suppressors of Beta-Amyloid has the potential to halt and regress disease progression [4].
Unlike traditional loss of function assays, which can only identify if a gene is a suppressor or not, using our more precise, dosage dependent optogenetic methods, we can characterize suppressor dynamics. The use of our optogenetic system can allow us to identify weak suppressors of beta-amyloid by screening for suppressors that yield median levels of beta-amyloid fluorescence. We have already identified the yeast ortholog of Beta-Amyloid by using the NIH BLASTp web service. We can conceivably begin screening for suppressors of Beta-Amyloid in the coming weeks when we acquire our transformation plasmid and a yeast gene deletion set. The platform we have developed in this work is not limited to Alzheimer’s disease, however; and will be used for identification of gene therapy targets for as varied conditions as Huntington’s disease, cancer, and diabetes [2,3].
Conclusion
In this work, we described the development, characterization, and optimization of an optogenetic toolkit for use in medical genetic studies. We were able to make improvements in the speed, cost, and accuracy of genetic screening by integrating yeast flow cytometric analysis with optogenetic control of transcription. We optimized and selected for three different fluorescent proteins that produced reliable levels of feedback for use in our toolkit and characterized our optogenetic system. We found that our optogenetic system produced equally reliable levels of gene induction as the widely-used GAL1 promoter, with a 22.0 second response time. To complement our experimental utlities, we utilized the modeling software TinkerCell to optimize values for numerous experimental variables in our system. Finally, we developed a C++ software program to take flow cytometry output from our system and process the data, streamlining the process of data analysis that limits current high-throughput techniques. Our toolkit was able to make a 5900-fold improvement in the speed of gene activation in medical genetic assays and a 7800-fold increase in the speed of analysis of flow cytometric data. Our experimental methods also yielded an 85.2% decrease in expenditures as compared to the use of mammalian cell lines. In the coming weeks, we will develop a custom yeast gene deletion set and loss of function assay as a proof-of-concept of our system, and begin testing an application of our toolkit by screening for orthologous suppressors of beta-amyloid that may be used in genetic therapy of Alzheimer’s disease. The construction of our toolkit will prompt advances in gene therapy and is a major step towards personalized therapy of a variety of diseases.
References
[1] S. Goverdhana, M. Puntel, W. Xiong, J. M. Zirger, C. Barcia, J. F. Curtin, E. B. Soffer, S. Mondkar, G. D. King, J. Hu, S. a Sciascia, M. Candolfi, D. S. Greengold, P. R. Lowenstein, and M. G. Castro, Molecular Therapy : the Journal of the American Society of Gene Therapy 12, 189 (2005).
[2] V. N. Ngo, R. E. Davis, L. Lamy, X. Yu, H. Zhao, G. Lenz, L. T. Lam, S. Dave, L. Yang, J. Powell, and L. M. Staudt, Nature 441, 106 (2006).
[3] W. Farris, S. Mansourian, M. a. Leissring, E. a. Eckman, L. Bertram, C. B. Eckman, R. E. Tanzi, and D. J. Selkoe, The American Journal of Pathology 164, 1425 (2004).
[4] J. J. Palop and L. Mucke, Nature Neuroscience 13, 812 (2010).
[5] J. Chen, M. B. Carter, B. S. Edwards, H. Cai, and L. a Sklar, Cytometry. Part A : the Journal of the International Society for Analytical Cytology 81, 90 (2012).
[6] L. Fenno, O. Yizhar, and K. Deisseroth, Annual Review of Neuroscience 34, 389 (2011).
[7] J. J. Tabor, A. Levskaya, and C. a Voigt, Journal of Molecular Biology 405, 315 (2011).
[8] L. Gardner and A. Deiters, Current Opinion in Chemical Biology 16, 292 (2012).
[9] F. Sherman, Methods Enzymol 350, (1991).
[10] J. Toettcher, C. Voigt, O. Weiner, and W. Lim, Nature Methods 8, 35 (2010).
[11] C. Mateus and S. V. Avery, Yeast (Chichester, England) 16, 1313 (2000).
[12] M. Manning and L. Hudgins, Genetics in Medicine : Official Journal of the American College of Medical Genetics 12, 742 (2010).
[13] D. Lohr, P. Venkov, and J. Zlatanova, The FASEB Journal 777 (1995).
[14] H. Ma, S. Kunes, P. Schatz, and D. Botstein, Gene 201 (1987).
[15] D. B. Hall and K. Struhl, The Journal of Biological Chemistry 277, 46043 (2002).
[16] H. Liu, X. Yu, K. Li, and J. Klejnot, Science … 322, 1535 (2008).
[17] C. Viappiani, S. Nonell, and M. Andreas, (2010).
[18] R. D. Gietz and R. a Woods, Methods in Enzymology 350, 87 (2002). Page 20
[19] C. Janke, M. M. Magiera, N. Rathfelder, C. Taxis, S. Reber, H. Maekawa, A. Moreno-Borchart, G. Doenges, E. Schwob, E. Schiebel, and M. Knop, Yeast (Chichester, England) 21, 947 (2004).
[20] C. D. Smolke, Nature Biotechnology 27, 1099 (2009).
[21] D. T. Gillespie, Annual Review of Physical Chemistry 58, 35 (2007).
[22] D. Chandran, F. T. Bergmann, and H. M. Sauro, Journal of Biological Engineering 3, 19 (2009).
[23] M. a Naivar, M. E. Wilder, R. C. Habbersett, T. a Woods, D. S. Sebba, J. P. Nolan, and S. W. Graves, Cytometry. Part A : the Journal of the International Society for Analytical Cytology 75, 979 (2009).
[24] C. B. Brachmann, a Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke, Yeast (Chichester, England) 14, 115 (1998).
[25] F. Giorgini, P. Guidetti, Q. Nguyen, S. C. Bennett, and P. J. Muchowski, Nature Genetics 37, 526 (2005).
[26] E. J. Olson and J. J. Tabor, Current Opinion in Chemical Biology 16, 300 (2012).
[27] R. M. Johnson, C. Allen, S. D. Melman, A. Waller, S. M. Young, L. a Sklar, and K. J. Parra, Analytical Biochemistry 398, 203 (2010).
[28] G. Charvin, F. R. Cross, and E. D. Siggia, PloS One 3, e1468 (2008).
[29] N. E. Buchler and F. R. Cross, Molecular Systems Biology 5, 272 (2009).
[30] S. Shimizu-Sato, E. Huq, J. M. Tepperman, and P. H. Quail, Nature Biotechnology 20, 1041 (2002).
 "
"