Team:Penn/Notebook/Biofilms
From 2012.igem.org
(Difference between revisions)
(→7/16/12) |
(→7/16/12) |
||
| Line 132: | Line 132: | ||
*pET-26b-luxS #1/#2 are slightly larger, indicating successful ligation, not as sure for pET-26b-luxS #3 | *pET-26b-luxS #1/#2 are slightly larger, indicating successful ligation, not as sure for pET-26b-luxS #3 | ||
*Last lane is unligated pET-26b vector | *Last lane is unligated pET-26b vector | ||
| - | [[File:pET-26b-luxSnocut071712.JPG|thumb|120px|alt=SCIENCE.|Image of 1% Agarose Gel of Miniprep products of pET-26b-luxS from three colonies]] | + | [[File:pET-26b-luxSnocut071712.JPG|thumb|120px|northeast|alt=SCIENCE.|Image of 1% Agarose Gel of Miniprep products of pET-26b-luxS from three colonies]] |
==7/17/12== | ==7/17/12== | ||
Latest revision as of 19:00, 18 July 2012
Contents |
6/6/12
- Set up equipment
- Autoclaved 2x 1L Sterile ddH2O
- Autoclaved 500 mL LB Broth for scale up of luxS (Plate 3 Well 2H, pSB1A2), plsr (Plate 2 Well 14H in pSB1A2 resistance to Amp)
- Added iGEM logo to wiki
6/7/12
- Resuspended available biobricks luxS & plsr (resuspended in 10uL ddH2O)
- Resuspended DNA transformed into DH5α (40uL DH5α+2uL DNA) & plated on LB plates w/ 15 uL of 100 mg/mL Amp spread on surface
- Requested lsrR & lsrK from iGEM HQ
6/8/12
- iGEM Wiki now split into two separate Notebooks
- Plenty of colonies, too many to count(TMTC) grew from plsr & luxS transformation from 6/7/12
- Selected one colony each
- Grew in 10 mL LB+Amp (100 ug/mL)
- Contacted researchers for V. harveyi bioassay & lysostaphin, V. harveyi must be purchased through ATCC ($300), lysostaphin obtained thru MTA.
6/9/12
- ~2mL of LB evaporated during incubation
- Cells spun down @ 5000 rpm for 10 min
- Miniprepped 4x Lysis into 1 column
- [plsr]=112 ng/uL
- [luxS]=114 ng/uL
6/11/12-6/16/12
- Dry lab work
6/18/12
- Obtained & resuspended primers for eGFP & plsr
6/19/12
- performed PCR w/ NEB standard taq kit w/ primers for eGFP & plsr
- Tested 2 annealing temperatures, 55C and 50C
- Ran gel w/ PCR product (2% agarose), saw clear eGFP bands, but possible .1kb obscured by loading dye.
6/20/12
- Performed qiagen PCR purification & nanodropped (it like it's hot)
6/21/12
- Ran gel of yesterday's PCR purification
- EtBr Gel 2% Agarose
- Lane 1: NEB 2-log ladder (1 ug loaded)
- Lane 2: PCR of eGFP using plsr_XhoI Fwd & eGFP_EcoRI Rev primers w/ 55C annealing temp.
- Lane 3: PCR of plsr using plsr_BglII Fwd & plsr_XhoI Rev primers w/ 55C annealing temp.
- Lane 4: PCR of eGFP using plsr_XhoI Fwd & eGFP_EcoRI Rev primers w/ 50C annealing temp.
- Lane 5: PCR of plsr using plsr_BglII Fwd & plsr_XhoI Rev primers w/ 50C annealing temp.
- Lane6/7: Null
- Lane 8: 1kb ladder left at rt o/n
- EtBr Gel 2% Agarose
- Obtained & transformed pET-26b
6/22/12
- Miniprepped pet26b and digested with BglII and EcorR
6/25/12
- Ran gel of eGFP/plsr ligation
- Ordered restriction enzymes from the Cell Center
6/26/12
- Attempted Gel Purification of digest products from 2/25/12
- Yields for Gel purification were too low to be useful (>1 ng/uL)
- Digested purified PCR rxns (annealing temperature=55C)
6/27/12
Ran gel of digested PCR rxns (55C), found no evidence of successful digestion.
6/28/12
Purified digest and attempted
6/29/12
- Performed digest of pET-26b, eGFP, and plsr
- Spread Biobrick shipment on Amp plates (lsrR, lsrK)
7/2/12
- Selected colonies from lsrK and lsrR streaked plates
- Grew in 10mL TB Broth
7/3/12
- Ran gel of ligated product, no size shift, unlikely that ligation was successful.
- Ligation rxn transformed to make sure
7/5/12
- Ligation transformation of pET-26-plsr-GFP failed, no colonies present
- Talked to Dan:
- When using primers to introduce restriction sites into
7/6/12
- Redesigned primers for plsr & egfp
- Set up TC Incubator & hood
- Contacted people in the FDA thru personal contacts for human practices
7/9/12
- Made Inactivating Solution for restriction digest
- 50mM EDTA (pH=8.0)
- 50% Glycerol
- Planned test restriction digest pET-26b troubleshooting tomorrow
- Trained new users for Tissue Culture
7/10/12
- Troubleshooting of pET-26 digestion
- Digest of pET-26b w/ BglII and XhoI
- Time course:
- 10 min
- 30 min
- 3 hr
- 6 hr
- o/n
- Inactivate w/ Inactivating Solution (10uL/50uL of restriction reaction)
- Nikita, Ashwin, Avin all have acceptable sterile practice
- Resuspended IDT luxS synthesized DNA
- Transformed synthesized luxS into DH5a
7/11/12
- Ran 2% gel of digest, did not see signs of any restriction
- Re-planned pET-26b digest troubleshooting
- Selected luxS colonies
7/12/12
- Thawed out 293T-CMV-Gaussia cells from Sarkar LN2
- Miniprepped luxS colony, digested w/ BamHI-HF & EcoRI-HF
- Performed gel extraction of luxS fragment
7/13/12
- Passaged 293T-CMV-Gaussia cells 1:10 & 1:5 for weekend
- Ligation of luxS w/ pET-26b
- Ligation rxn transformed into DH5a
7/14/12
- Ligation showed successful transformants
7/15/12
- Colonies selected by Mike, grown in 10mL LB
7/16/12
- SAAST Presentation
- Miniprep of 10mL pET-26b-luxS cultures
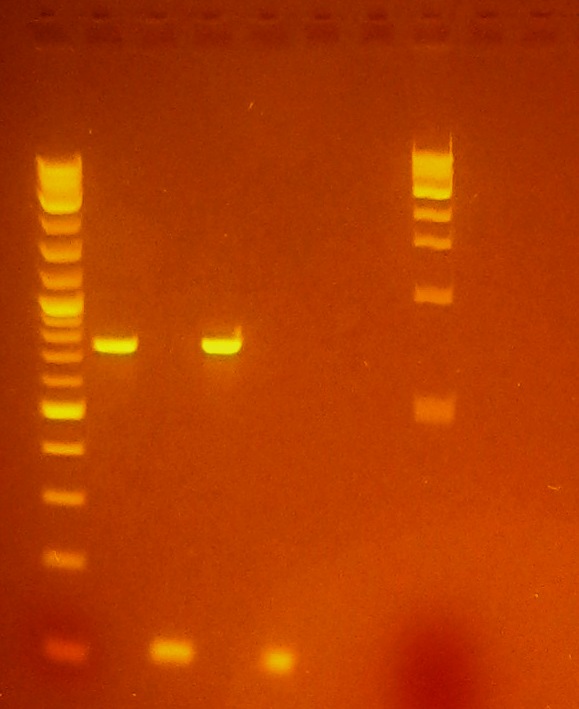
- Ran gel of minipreps from 3 pET-26b-luxS colonies to confirm successful ligation
- pET-26b-luxS #1/#2 are slightly larger, indicating successful ligation, not as sure for pET-26b-luxS #3
- Last lane is unligated pET-26b vector
7/17/12
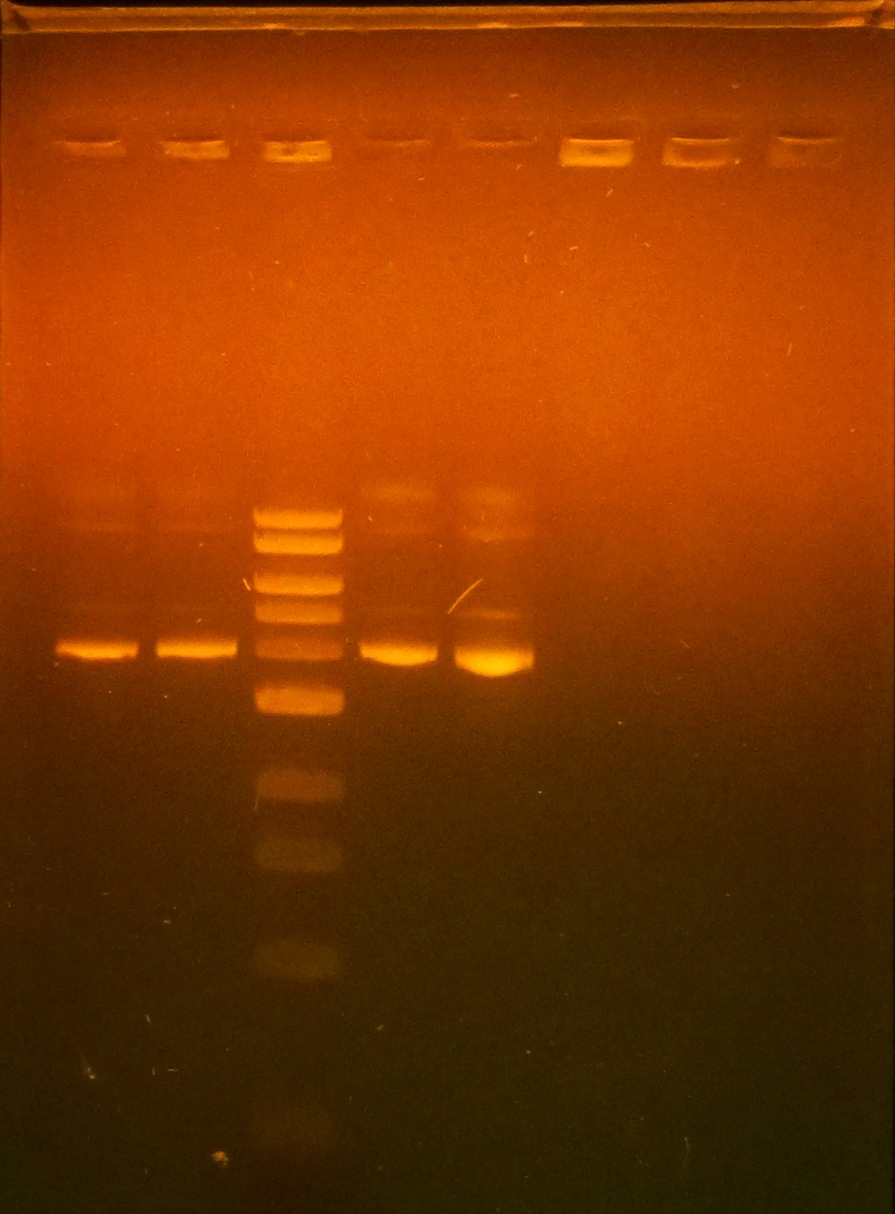
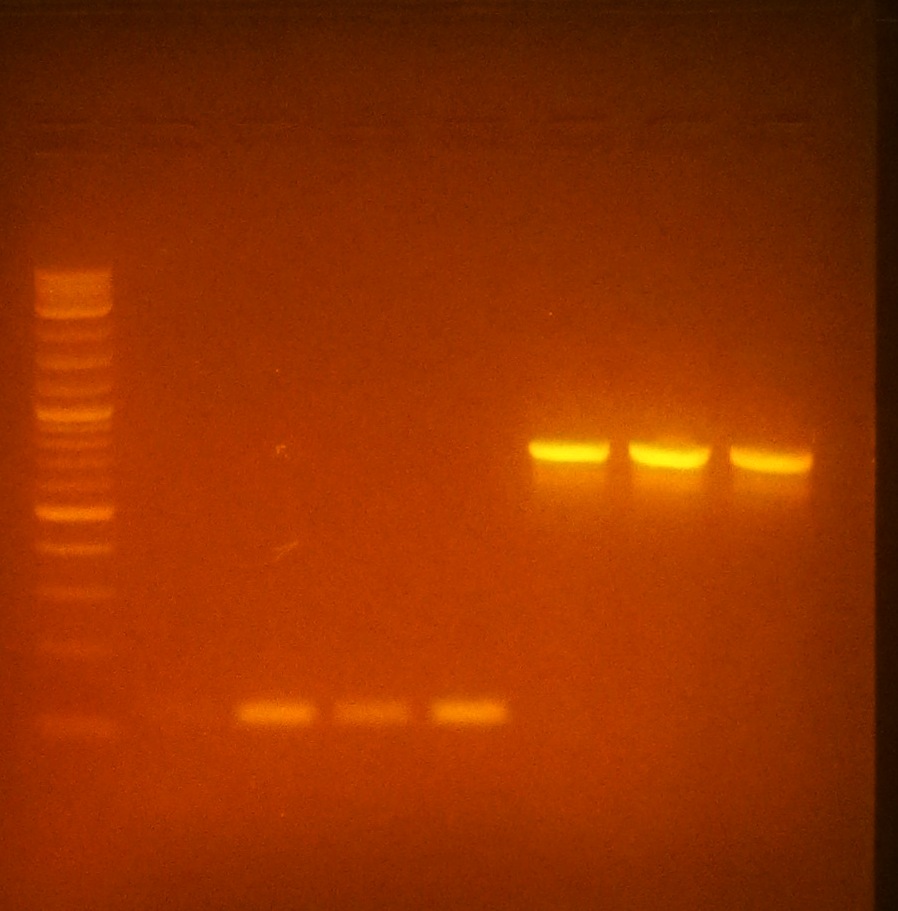
- Ran gel to determine if insertion of pET-26b-luxS was correctly ligated
- Gel showed there was a possible size shift corresponding to insertion in all three selected colonies of pET-26b-luxS colonies
- From left to right (optimum temperatures bolded): Ladder, plsr @45C,52C,58C,65C, egfp @52C,58C,65C
- Performed 100uL PCR of plsr @65 & 100uL PCR of egfp @58 for further ligation experiments.
 "
"