Team:Colombia/Project/Experiments/Pseudomonas
From 2012.igem.org
Sylvita1015 (Talk | contribs) (→Pseudomonas) |
Sylvita1015 (Talk | contribs) (→Toxin-Antitoxin) |
||
| (50 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{https://2012.igem.org/User:Tabima}} | |
| - | < | + | <div class="right_box"> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
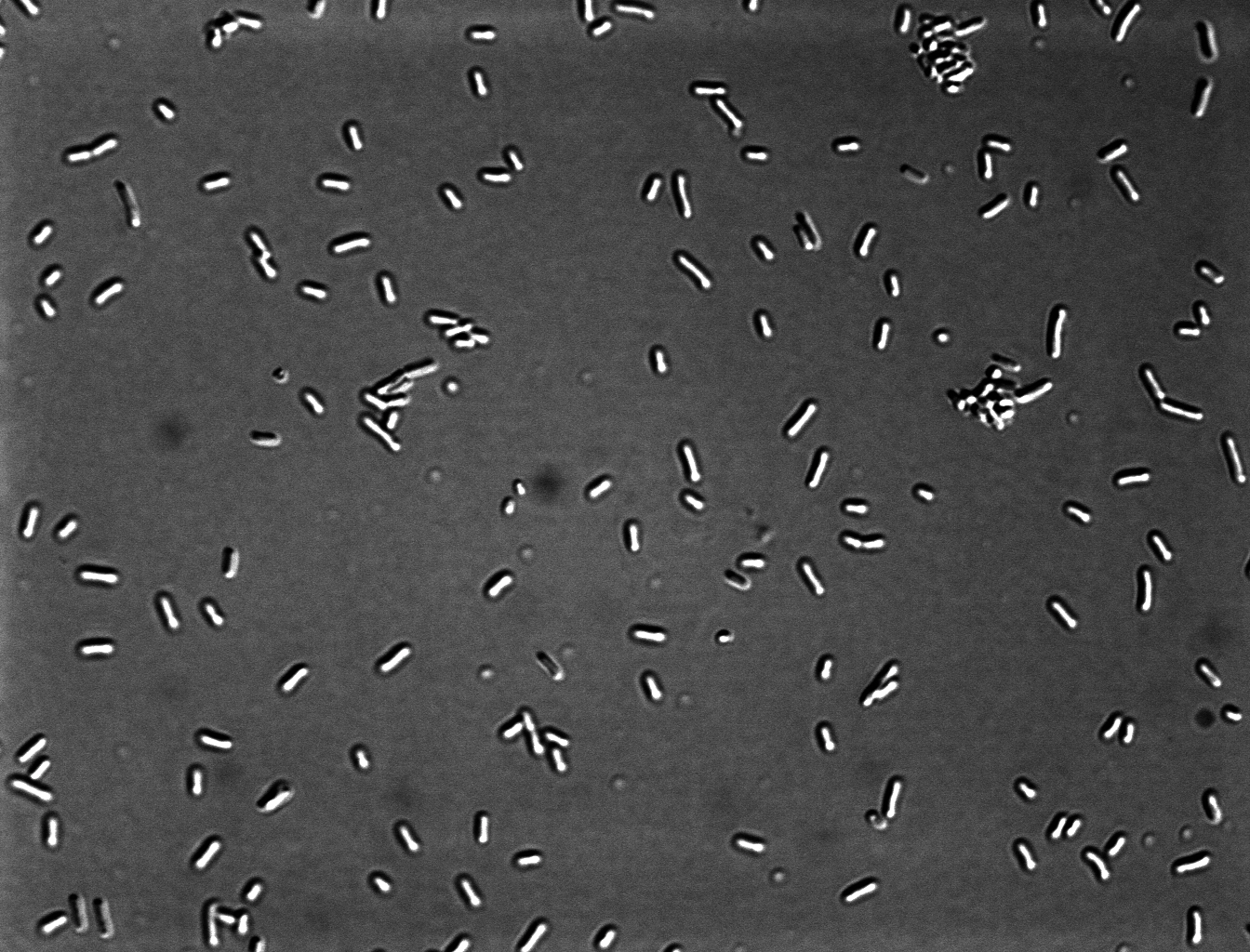
| - | + | =''' ''Escherichia coli'' K12 '''= | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[File:DICx60_E_coli.jpg|center|650 px]] | |
| - | + | ||
| - | + | ||
| - | + | = ''' Toxin-Antitoxin ''' = | |
| - | + | ||
| - | + | ||
| - | . | + | We propose to use Toxin-Antitoxin (TA) chromosomal modules of ''Escherichia coli'' to induce type I persistency, a physiological state characterized by the arrested growth of bacterial cells [Lewis et al, 2010], for population control of GMOs and to prevent both HGT and plasmid curing under environmental conditions without antibiotic pressures. |
| - | + | ||
| - | + | ||
| - | + | <html> | |
| - | + | <br> | |
| - | + | <center> | |
| + | <iframe width="700" height="500" src="http://youtu.be/j8Ya4r4dWy0" frameborder="0" allowfullscreen></iframe> | ||
| + | </center> | ||
| + | </br> | ||
| + | </html> | ||
| - | + | As can be seen in the [http://youtu.be/j8Ya4r4dWy0 video], type I persister cells from the high persistence ''E. coli'' mutant strain TH1269 (''hipA7'') remain non growing for several hours, and at a random moment they “wake up” and start to grow rapidly. | |
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
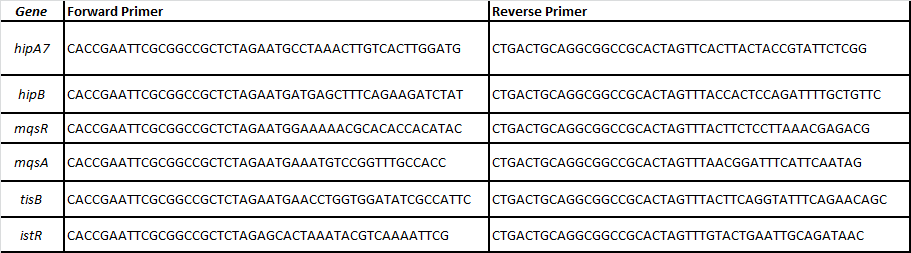
| - | + | For that, we designed specific primers for each ORF of the following TA pairs related with persistence (Balaban et al, 2004; Lewis 2010; Maisonneuve et al, 2011): | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | -HipA7/HipB | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | -MqsR/MqsA | |
| - | / | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | -TisB/istR | |
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | [[File:PrimersTAiGEM.png|center|700px]] | |
| - | </ | + | |
| + | |||
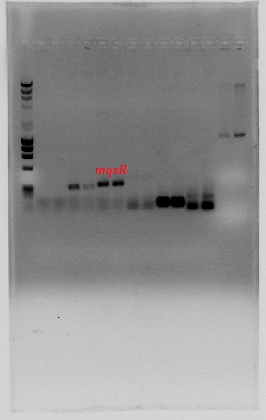
| + | In order to enable other teams to use said toxin-antitoxin modules we cloned these parts without promoters in chloramphenicol backbones. '''So far we have constructed the BioBricks with the toxins [http://partsregistry.org/Part:BBa_K831000:Design HipA7] and [http://partsregistry.org/Part:BBa_K831003:Design TisB], and with the antitoxins [http://partsregistry.org/Part:BBa_K831001:Design HipB], [http://partsregistry.org/Part:BBa_K831002:Design MqsA] and [http://partsregistry.org/Part:BBa_K831004:Design ''istR'' ].''' | ||
| + | |||
| + | |||
| + | [[File:Confirmacion TA Cloramfenicol.png|center|300px]] | ||
| + | |||
| + | |||
| + | |||
| + | We were going to use the ''wild type'' promoters of the Toxin-Antitoxin modules; however the deterministic model gave us feedback which established that inducible promoters were much more likely to give us the results we were looking for. | ||
| + | |||
| + | For the construction of our inducible parts, we first tried to build the toxins under the control of the Lac promoter and the antitoxins under the tetR promoter. '''We succeeded in the construction of the parts containing the [http://partsregistry.org/Part:BBa_K831009 plac-''hipB''] and [http://partsregistry.org/Part:BBa_K831014:Design plac-''istR''] antitoxins and the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K831007 ptetR-''mqsR''] inducible toxin'''. | ||
| + | |||
| + | [[File:MqsR2.jpg|center|300px]] | ||
| + | |||
| + | But due to the leakage of lac promoter and even that of the tetR promoter (has less leakage that lac) we were unable to construct the remaining toxin parts using both of those promoters. To overcome this inconvenience we decided to use the prm promoter which is inducible by the CI lambda phage protein. '''So far we have constructed the inducible [http://partsregistry.org/Part:BBa_K831015 prm-hipA7] BioBrick'''. | ||
| + | |||
| + | In order to assess the functionality of these parts, we cloned ''hipB'' under the control of lac promoter in an ''Escherichia coli'' K12 strain that contains hipA7 in its chromosome (''E. coli'' K12 MG1655 TH1269). | ||
| + | |||
| + | As HipB neutralizes the induction of persistency caused by HipA7 in this strain, we induced our strain with IPTG at different concentrations and evaluate the persisters frequencies implementing an original protocol for persister cells isolation based on lysis (S. Cañas et al., Manuscript in preparation). The results of this caractherization can be seen in the main page of [http://partsregistry.org/Part:BBa_K831009 plac-''hipB''], BioBrick BBa_K831009. | ||
| + | |||
| + | We constructed inducible CI under the control of the lac promoter in order to analyze the effect of the induction of HipA7 in the persisters frequency on a wild type ''E. coli'' K12 strain. The results of this caractherization can be seen in the main page of [http://partsregistry.org/Part:BBa_K831015 prm-''hipA7''], BioBrick BBa_K831015. | ||
| + | |||
| + | =''' Assays in ''Pseudomonas fluorescens'' '''= | ||
| + | |||
| + | In order to improve the stability and survival of our system we are assessing the possibility of using ''P. fluorescens'' as our final chassis. For this purpose, we first standardized the normal recount of persisters of a ''P. fluorescens wild type'' strain without induction. We used an original protocol for determining persistence based on lysis from work that is not part of iGEM (S. Cañas et al., Manuscript in preparation), which we validated for use in ''P. fluorescens''. | ||
| + | |||
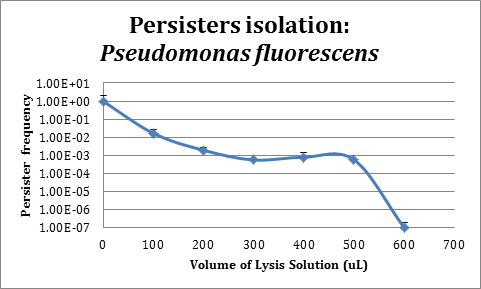
| + | [[File:Persistence_Pseudomonas_iGEM.png|center|500px]] | ||
| + | |||
| + | As can be seen in the figure, the protocol implemented for persister isolation works properly in ''P. fluorescens'' and will allow us to further test in this strain if population control using persistency induction by the TA module from ''Escherichia coli'' K12 HipA7/HipB could be implemented. | ||
| + | |||
| + | |||
| + | =''' References'''= | ||
| + | |||
| + | '''1.'''Balaban N., et al. Bacterial persistence as a phenotypic switch. 2004. Science 305:1622. | ||
| + | |||
| + | '''2.'''Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357-372. | ||
| + | |||
| + | '''3.'''Maisonneuve, E,. Shakespeare, L J,.Jørgensen, M G,.Gerdes, K,. Bacterial persistence by RNA endonucleases. 2011.Proceedings of theNationalAcademy of Sciences. 10.1073/pnas.1100186108 | ||
| + | |||
| + | </div> | ||
Latest revision as of 03:24, 27 October 2012
Template:Https://2012.igem.org/User:Tabima
Contents |
Escherichia coli K12
Toxin-Antitoxin
We propose to use Toxin-Antitoxin (TA) chromosomal modules of Escherichia coli to induce type I persistency, a physiological state characterized by the arrested growth of bacterial cells [Lewis et al, 2010], for population control of GMOs and to prevent both HGT and plasmid curing under environmental conditions without antibiotic pressures.
As can be seen in the [http://youtu.be/j8Ya4r4dWy0 video], type I persister cells from the high persistence E. coli mutant strain TH1269 (hipA7) remain non growing for several hours, and at a random moment they “wake up” and start to grow rapidly.
For that, we designed specific primers for each ORF of the following TA pairs related with persistence (Balaban et al, 2004; Lewis 2010; Maisonneuve et al, 2011):
-HipA7/HipB
-MqsR/MqsA
-TisB/istR
In order to enable other teams to use said toxin-antitoxin modules we cloned these parts without promoters in chloramphenicol backbones. So far we have constructed the BioBricks with the toxins [http://partsregistry.org/Part:BBa_K831000:Design HipA7] and [http://partsregistry.org/Part:BBa_K831003:Design TisB], and with the antitoxins [http://partsregistry.org/Part:BBa_K831001:Design HipB], [http://partsregistry.org/Part:BBa_K831002:Design MqsA] and [http://partsregistry.org/Part:BBa_K831004:Design istR ].
We were going to use the wild type promoters of the Toxin-Antitoxin modules; however the deterministic model gave us feedback which established that inducible promoters were much more likely to give us the results we were looking for.
For the construction of our inducible parts, we first tried to build the toxins under the control of the Lac promoter and the antitoxins under the tetR promoter. We succeeded in the construction of the parts containing the [http://partsregistry.org/Part:BBa_K831009 plac-hipB] and [http://partsregistry.org/Part:BBa_K831014:Design plac-istR] antitoxins and the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K831007 ptetR-mqsR] inducible toxin.
But due to the leakage of lac promoter and even that of the tetR promoter (has less leakage that lac) we were unable to construct the remaining toxin parts using both of those promoters. To overcome this inconvenience we decided to use the prm promoter which is inducible by the CI lambda phage protein. So far we have constructed the inducible [http://partsregistry.org/Part:BBa_K831015 prm-hipA7] BioBrick.
In order to assess the functionality of these parts, we cloned hipB under the control of lac promoter in an Escherichia coli K12 strain that contains hipA7 in its chromosome (E. coli K12 MG1655 TH1269).
As HipB neutralizes the induction of persistency caused by HipA7 in this strain, we induced our strain with IPTG at different concentrations and evaluate the persisters frequencies implementing an original protocol for persister cells isolation based on lysis (S. Cañas et al., Manuscript in preparation). The results of this caractherization can be seen in the main page of [http://partsregistry.org/Part:BBa_K831009 plac-hipB], BioBrick BBa_K831009.
We constructed inducible CI under the control of the lac promoter in order to analyze the effect of the induction of HipA7 in the persisters frequency on a wild type E. coli K12 strain. The results of this caractherization can be seen in the main page of [http://partsregistry.org/Part:BBa_K831015 prm-hipA7], BioBrick BBa_K831015.
Assays in Pseudomonas fluorescens
In order to improve the stability and survival of our system we are assessing the possibility of using P. fluorescens as our final chassis. For this purpose, we first standardized the normal recount of persisters of a P. fluorescens wild type strain without induction. We used an original protocol for determining persistence based on lysis from work that is not part of iGEM (S. Cañas et al., Manuscript in preparation), which we validated for use in P. fluorescens.
As can be seen in the figure, the protocol implemented for persister isolation works properly in P. fluorescens and will allow us to further test in this strain if population control using persistency induction by the TA module from Escherichia coli K12 HipA7/HipB could be implemented.
References
1.Balaban N., et al. Bacterial persistence as a phenotypic switch. 2004. Science 305:1622.
2.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357-372.
3.Maisonneuve, E,. Shakespeare, L J,.Jørgensen, M G,.Gerdes, K,. Bacterial persistence by RNA endonucleases. 2011.Proceedings of theNationalAcademy of Sciences. 10.1073/pnas.1100186108
 "
"