Team:Cornell/Notebook/Salicylate reporter/July8
From 2012.igem.org
(→July 10th: added picture of new freezer) |
(→July 8th-14th) |
||
| (22 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
=July 8th-14th= | =July 8th-14th= | ||
| + | The focus of this week was to transfer our arsenic sensing plasmids to Shewanella strain JG700 via congugation with E.coli strain WM3064. After struggling to electroporate control plasmids, we consulted with Dr. Gralnick who reccomended proceeding with conjugation and kindly provided E.coli strain WM3064. This strain is auxotrophic for 2,6-Diaminopimelic Acid (DAP), which makes the strain useful for selecting against non-Shewanella conjugants. | ||
| + | |||
==July 8th== | ==July 8th== | ||
| Line 10: | Line 12: | ||
==July 9th== | ==July 9th== | ||
[[Image:2012_7_9_FearlessLeaderAkaLordOfFlame.jpg|thumb|right|Claire spark-lighting a Bunsen burner.]] | [[Image:2012_7_9_FearlessLeaderAkaLordOfFlame.jpg|thumb|right|Claire spark-lighting a Bunsen burner.]] | ||
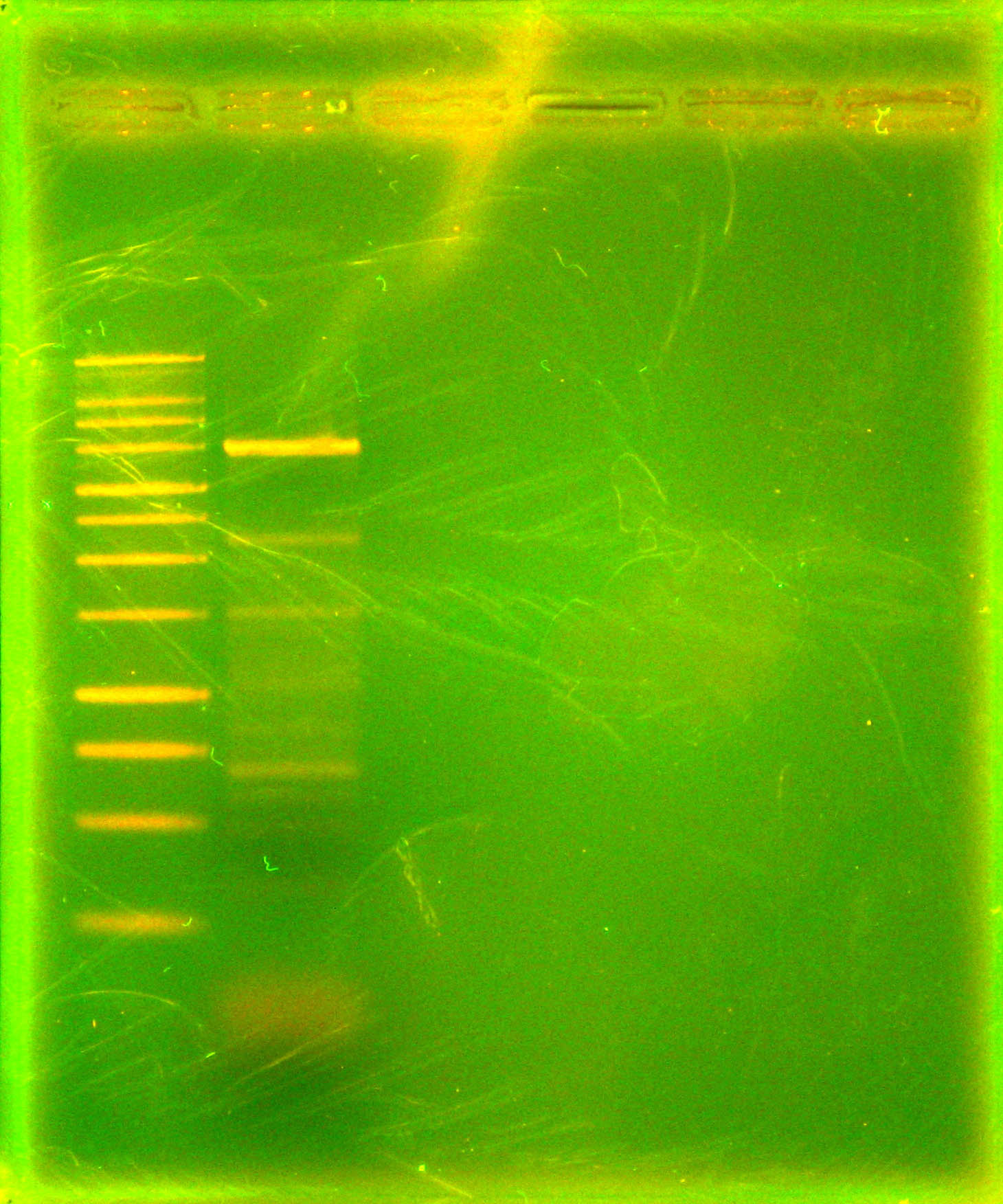
| - | Dylan performed a [[Team:Cornell/Notebook/Phusion_PCR|Phusion PCR]] with a single annealing temperature of 59 degrees Celsius using primers 27 and 28 for the final time. In the future, new primers will be used. The purpose of this PCR was to amplify [[Team:Cornell/Notebook/StrainList| | + | Dylan performed a [[Team:Cornell/Notebook/Phusion_PCR|Phusion PCR]] with a single annealing temperature of 59 degrees Celsius using primers 27 and 28 for the final time. In the future, new primers will be used. The purpose of this PCR was to amplify p21 (See: [[Team:Cornell/Notebook/StrainList|strain list]]). The PCR product was analyzed using [[Team:Cornell/Notebook/Gel_electrophoresis|agarose gel electrophoresis]] to determine if the desired 1.127 kb fragment is present. |
| - | To transform our plasmids into Shewanella with conjugation, we had asked Dr. Jeff Gralnick for WM3064 E. coli for which plates containing 2,6-Diaminopimelic Acid (DAP) are required. In preparation to receive this strain, the plates with DAP were prepared. | + | To transform our plasmids into Shewanella with conjugation, we had asked Dr. Jeff Gralnick for WM3064 E. coli for which plates containing 2,6-Diaminopimelic Acid (DAP) are required. In preparation to receive this strain, the plates with DAP and DAP/kanamycin were prepared. |
| - | + | ||
==July 10th== | ==July 10th== | ||
| - | Caleb [[Team:Cornell/Notebook/Miniprep|miniprepped]] strains containing plasmids p8, p9, p10, p15, and p16 (See | + | [[Image:2012_7_10_DylanNewFridge.jpg|thumb|left|Dylan setting up our new freezer!]]Caleb [[Team:Cornell/Notebook/Miniprep|miniprepped]] strains containing plasmids p8, p9, p10, p15, and p16 (See: [[Team:Cornell/Notebook/StrainList|strain list]]) Also, our new freezer arrived. We cleaned and welcomed our new friend. |
| - | + | The WM3064 came in and we plated from the agar stab. Dylan set up two 18 mL/hr continuous flow reactors using 10x diluted LB and innoculated one of the reactors with JG700 and one with wild type Shewanella oneidensis MR-1. | |
==July 11th== | ==July 11th== | ||
| + | Dylan started a WM3064 subculture in the morning before meeting Swati, Shweta, Caleb, and Danielle at the Boyce Thompson Institute for Plant Research to give a presentation both on our project and on the broader applications of synthetic biology to the energy industry to a group of high school teachers from New York state. After the presentation, Dylan and Danielle prepared plates containing DAP/chloramphenicol and DAP/ampicillin to grow transformed WM3064 on, while Mark continued preparing [[electrocompetent stocks]] of WM3064 with the subculture that Dylan had started. | ||
| + | |||
| + | We transformed things later that day. | ||
==July 12th== | ==July 12th== | ||
| + | [[Image:2012_7_12_p21FailBlog.jpg|thumb|right|Gel showing failed p21 PCR product]] | ||
| + | Shweta prepared a glycerol stock of WM3064 for future use, and Dylan plated some remaining transformed WM3064 hoping to get more colonies of the successfully transformed cells. | ||
| + | Dylan and Mark submitted p8, p9, and p10 for sequencing using two different primers for each plasmid. | ||
| + | |||
| + | The previous night's p21 PCR seemed to have failed, even with the new primers. Based on the [[Team:Cornell/Notebook/Gel_electrophoresis|gel]] run by Claire, then analyzed by Dylan and Caleb. | ||
==July 13th== | ==July 13th== | ||
| + | [[Image:2012_7_13_DylanConjugating.jpg|thumb|left|Dylan plating putatively promiscuous E. coli mixed with S. oneidensis.]] | ||
| + | |||
| + | Today began with Dylan and Caleb setting up conjugations between our transformed donor E. coli and S. oneidensis. | ||
| + | |||
| + | The sequencing results for p8, p9 and p10 all came back as failed, so it was concluded that the Gibson assembly failed. There were five locations on the nah operon at which sequencing could begin, of which we considered all but the fourth. Sequencing at the first location resulted in a sequence that corresponded to the fifth sequencing location, with some sequence before it and all of the expected sequence after it. The fifth sequencing location produced the expected sequence. Then the middle two regions which we considered all resulted in failed sequencing. This has led us to the conclusion that segments that are required in the operon are missing, and thus the assembly failed. Dylan and Caleb began attempting to look into the possibility of this being a cause, and some potential solutions. | ||
| + | <!--- | ||
| + | Above still needs editing. | ||
| + | ---> | ||
| + | |||
| + | |||
| + | In discussing why p21 PCR seems to continue failing, the idea of having p21 sequenced arose. Dylan prepared the sample and submitted it for sequencing. Meanwhile, Mark set up yet another [[Team:Cornell/Notebook/Phusion_PCR|Phusion PCR]] of p21, this time with a higher annealing temperature of 61 degrees Celsius and a decreased extension time of 18 seconds. The goal was to prevent the mispriming which seemed to have occurred in the previous day's attempt. | ||
| + | Later that day, Dylan set up yet another PCR with a decreased annealing temperature of 55 degrees Celsius in order to empirically narrow-in on the optimal annealing temperature for the amplification of the salicylate sensing region from p21. | ||
| + | |||
| + | <!--- | ||
| + | Explain the next step of conjugation and what other PCR was performed. | ||
| + | ---> | ||
==July 14th== | ==July 14th== | ||
| + | Swati ran a gel of 5uL of the p21 PCR that Mark set up yesterday. After staining, the DNA ladder was well visualized but there were no bands in the p21 lane, suggesting that the PCR did not work. In a fit of rage, Swati yelled at Maneesh for going to get a drink of water - yet another innocent victim of failed PCR. | ||
Latest revision as of 04:08, 16 July 2012
| Home | Team | Project | Parts | Modeling | Notebook | Protocols | Safety | Attributions |
|---|
|
The salicylate reporter is analogous to the arsenic reporter, but with a salicylate-sensitive promoter. That is, in the presence of salicylate it will produce MtrB, completing the Mtr pathway and allowing Shewanella to produce current in our biosensor. In combination with the nah operon, this reporter will be able to detect naphthalene levels. Back to salicylate reporter week view
July 8th-14th
The focus of this week was to transfer our arsenic sensing plasmids to Shewanella strain JG700 via congugation with E.coli strain WM3064. After struggling to electroporate control plasmids, we consulted with Dr. Gralnick who reccomended proceeding with conjugation and kindly provided E.coli strain WM3064. This strain is auxotrophic for 2,6-Diaminopimelic Acid (DAP), which makes the strain useful for selecting against non-Shewanella conjugants.
July 8th
July 9th
Dylan performed a Phusion PCR with a single annealing temperature of 59 degrees Celsius using primers 27 and 28 for the final time. In the future, new primers will be used. The purpose of this PCR was to amplify p21 (See: strain list). The PCR product was analyzed using agarose gel electrophoresis to determine if the desired 1.127 kb fragment is present. To transform our plasmids into Shewanella with conjugation, we had asked Dr. Jeff Gralnick for WM3064 E. coli for which plates containing 2,6-Diaminopimelic Acid (DAP) are required. In preparation to receive this strain, the plates with DAP and DAP/kanamycin were prepared.
July 10th
Caleb miniprepped strains containing plasmids p8, p9, p10, p15, and p16 (See: strain list) Also, our new freezer arrived. We cleaned and welcomed our new friend.The WM3064 came in and we plated from the agar stab. Dylan set up two 18 mL/hr continuous flow reactors using 10x diluted LB and innoculated one of the reactors with JG700 and one with wild type Shewanella oneidensis MR-1.
July 11th
Dylan started a WM3064 subculture in the morning before meeting Swati, Shweta, Caleb, and Danielle at the Boyce Thompson Institute for Plant Research to give a presentation both on our project and on the broader applications of synthetic biology to the energy industry to a group of high school teachers from New York state. After the presentation, Dylan and Danielle prepared plates containing DAP/chloramphenicol and DAP/ampicillin to grow transformed WM3064 on, while Mark continued preparing electrocompetent stocks of WM3064 with the subculture that Dylan had started.
We transformed things later that day.
July 12th
Shweta prepared a glycerol stock of WM3064 for future use, and Dylan plated some remaining transformed WM3064 hoping to get more colonies of the successfully transformed cells.
Dylan and Mark submitted p8, p9, and p10 for sequencing using two different primers for each plasmid.
The previous night's p21 PCR seemed to have failed, even with the new primers. Based on the gel run by Claire, then analyzed by Dylan and Caleb.
July 13th
Today began with Dylan and Caleb setting up conjugations between our transformed donor E. coli and S. oneidensis.
The sequencing results for p8, p9 and p10 all came back as failed, so it was concluded that the Gibson assembly failed. There were five locations on the nah operon at which sequencing could begin, of which we considered all but the fourth. Sequencing at the first location resulted in a sequence that corresponded to the fifth sequencing location, with some sequence before it and all of the expected sequence after it. The fifth sequencing location produced the expected sequence. Then the middle two regions which we considered all resulted in failed sequencing. This has led us to the conclusion that segments that are required in the operon are missing, and thus the assembly failed. Dylan and Caleb began attempting to look into the possibility of this being a cause, and some potential solutions.
In discussing why p21 PCR seems to continue failing, the idea of having p21 sequenced arose. Dylan prepared the sample and submitted it for sequencing. Meanwhile, Mark set up yet another Phusion PCR of p21, this time with a higher annealing temperature of 61 degrees Celsius and a decreased extension time of 18 seconds. The goal was to prevent the mispriming which seemed to have occurred in the previous day's attempt.
Later that day, Dylan set up yet another PCR with a decreased annealing temperature of 55 degrees Celsius in order to empirically narrow-in on the optimal annealing temperature for the amplification of the salicylate sensing region from p21.
July 14th
Swati ran a gel of 5uL of the p21 PCR that Mark set up yesterday. After staining, the DNA ladder was well visualized but there were no bands in the p21 lane, suggesting that the PCR did not work. In a fit of rage, Swati yelled at Maneesh for going to get a drink of water - yet another innocent victim of failed PCR.
 "
"