Team:Cornell/Notebook/Salicylate reporter/July1
From 2012.igem.org
(→July 4th, Wednesday) |
(→July 1st, Sunday) |
||
| (24 intermediate revisions not shown) | |||
| Line 8: | Line 8: | ||
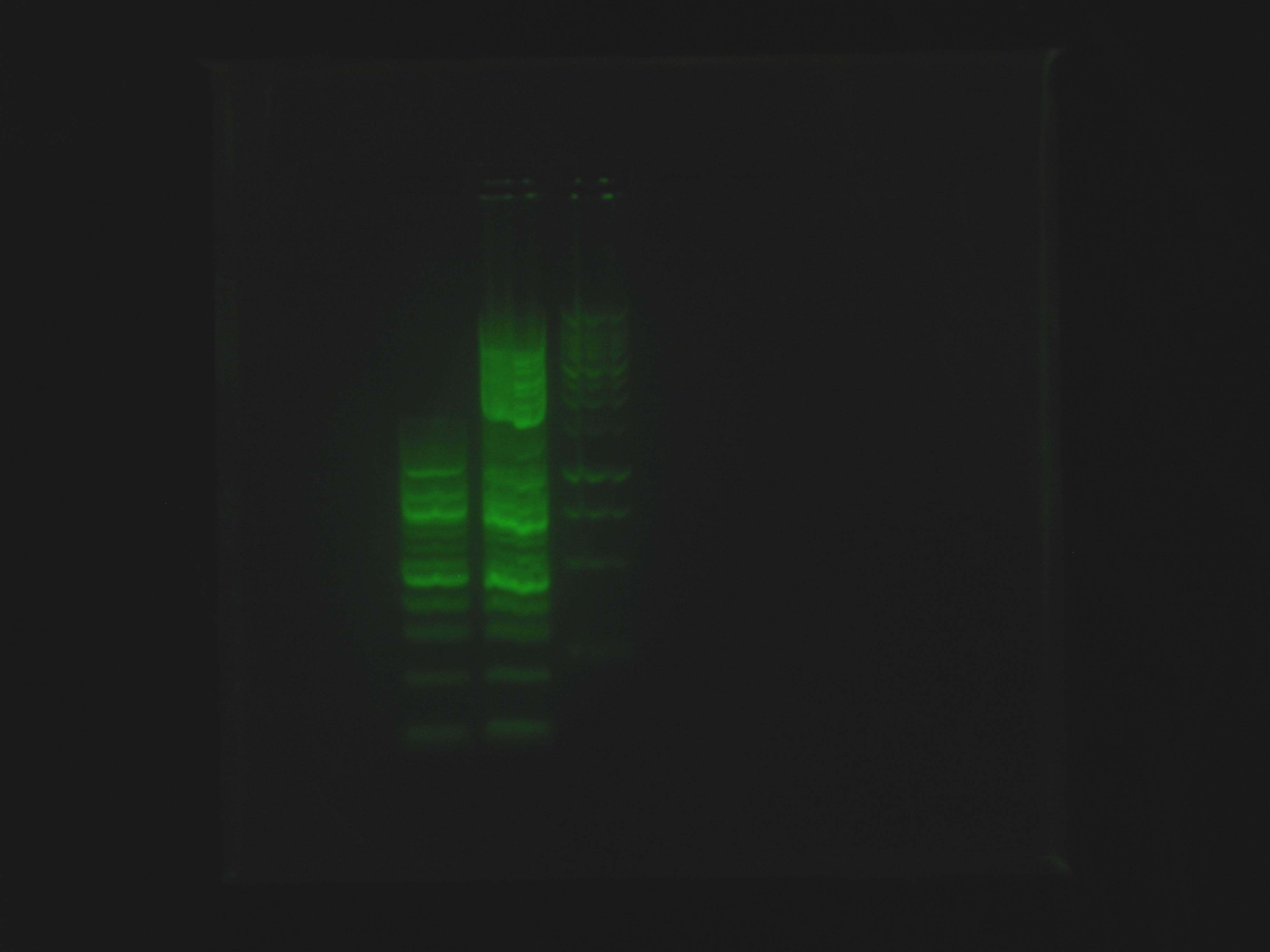
[[File:2012_07_01_LadderExperiment2.jpg|right|thumb|SYBR Green stained gel of Ladder Experiment]] | [[File:2012_07_01_LadderExperiment2.jpg|right|thumb|SYBR Green stained gel of Ladder Experiment]] | ||
At 9:50, Dylan and Caleb set up 2 gels for [[Team:Cornell/Notebook/Gel_electrophoresis|electrophoresis]]. Caleb's was 1% agarose in BIO-RAD Mini-Sub Cell system for continuation of ladder test using SYBR Green, containing NEB 100 bp ladder, NEB 2-log ladder, Promega 1 kb ladder and run at 100V. Dylan's was 1% agarose in Owl box using ethidium bromide, containing the nah operon PCR product from previous night, and run at 55 V. | At 9:50, Dylan and Caleb set up 2 gels for [[Team:Cornell/Notebook/Gel_electrophoresis|electrophoresis]]. Caleb's was 1% agarose in BIO-RAD Mini-Sub Cell system for continuation of ladder test using SYBR Green, containing NEB 100 bp ladder, NEB 2-log ladder, Promega 1 kb ladder and run at 100V. Dylan's was 1% agarose in Owl box using ethidium bromide, containing the nah operon PCR product from previous night, and run at 55 V. | ||
| + | |||
| + | Our plates of DH5alpha transformed with p21 ligated into pBBRBB with mtrB grew only one colony, possibly contamination. Dylan ran a colony PCR and got a smear, suggesting that the PCR of p21 or the ligation did not work. | ||
Because we learned that our SYBR Green was causing ladder to run strangely, Dylan decided to redo a [[Team:Cornell/Notebook/Vent_PCR|Vent PCR]] to amplify the salicylate reporter region out of p21. | Because we learned that our SYBR Green was causing ladder to run strangely, Dylan decided to redo a [[Team:Cornell/Notebook/Vent_PCR|Vent PCR]] to amplify the salicylate reporter region out of p21. | ||
| - | Also, a liquid culture of JG700 was prepared, as well as replating of p21, p22, JG700, JG1220, JG1537, JG1219. (See | + | Also, a liquid culture of JG700 was prepared, as well as replating of p21, p22, JG700, JG1220, JG1537, JG1219. (See: [[Team:Cornell/Notebook/StrainList|strain list]]) |
==July 2nd, Monday== | ==July 2nd, Monday== | ||
| Line 17: | Line 19: | ||
Today, Dylan and Caleb ran a [[Team:Cornell/Notebook/Gel_electrophoresis|gel]] of a [[Team:Cornell/Notebook/Vent_PCR|Vent PCR]] of p21 (the PCR product being the salicylate reporter) at 55 V. Additionally, Caleb decided to run a control gel at 100 V to determine whether higher voltage was a factor in our previous ladder problems (in addition to the SYBR Green stock). While we determined that the higher voltage did not cause our ladder issues, we did not see any bands from the p21 PCR. | Today, Dylan and Caleb ran a [[Team:Cornell/Notebook/Gel_electrophoresis|gel]] of a [[Team:Cornell/Notebook/Vent_PCR|Vent PCR]] of p21 (the PCR product being the salicylate reporter) at 55 V. Additionally, Caleb decided to run a control gel at 100 V to determine whether higher voltage was a factor in our previous ladder problems (in addition to the SYBR Green stock). While we determined that the higher voltage did not cause our ladder issues, we did not see any bands from the p21 PCR. | ||
| - | Dylan prepared electrocompetent cells using the Myers and Myers protocol. Modified protocol using 2 5mL cultures @ 4000g for 10 min. Washed with 2 mL sorbitol, resuspended in 100 uL sorbitol. First electroporation ts = 2.32 ms, second ts = 2.02 ms. Added 1 uL of plasmid (575.5 ng) to each cuvette. First used .60 V, then 0.55 V, both with R = 400 ohms. | + | Dylan prepared electrocompetent cells using the [[Team:Cornell/Notebook/Transformation_shewanella#Myers_and_Myers|Myers and Myers]] protocol. Modified protocol using 2 5mL cultures @ 4000g for 10 min. Washed with 2 mL sorbitol, resuspended in 100 uL sorbitol. First electroporation ts = 2.32 ms, second ts = 2.02 ms. Added 1 uL of plasmid (575.5 ng) to each cuvette. First used .60 V, then 0.55 V, both with R = 400 ohms. |
| - | Mark and Danielle started liquid cultures of S1, S9, S10, S11, S15, S16, S18, S27 (See: [[Team:Cornell/Notebook/StrainList| | + | Mark and Danielle started liquid cultures of S1, S9, S10, S11, S15, S16, S18, S27 (See: [[Team:Cornell/Notebook/StrainList|strain list]]) |
==July 3rd, Tuesday== | ==July 3rd, Tuesday== | ||
| - | Caleb [[Team:Cornell/Notebook/Miniprep|miniprepped]] S16 (p15), S22 (p21), S9 (p8), S10 (p9), S11 (p10), S18 (p17), and S15 (p14). Instead of a single EB elution, Caleb did two elutions, 30 ul each. The double 30 ul elution turned out to be effective at recovering a usable amount of DNA in the second elution, so we're including it on our miniprep protocol for future minipreps. | + | Caleb [[Team:Cornell/Notebook/Miniprep|miniprepped]] S16 (p15), S22 (p21), S9 (p8), S10 (p9), S11 (p10), S18 (p17), and S15 (p14). (See: [[Team:Cornell/Notebook/StrainList|strain list]]) Instead of a single EB elution, Caleb did two elutions, 30 ul each. The double 30 ul elution turned out to be effective at recovering a usable amount of DNA in the second elution, so we're including it on our miniprep protocol for future minipreps. |
<!-- | <!-- | ||
| Line 36: | Line 38: | ||
J = p14 | J = p14 | ||
K = p14 | K = p14 | ||
| - | |||
| - | |||
--> | --> | ||
| - | |||
| - | |||
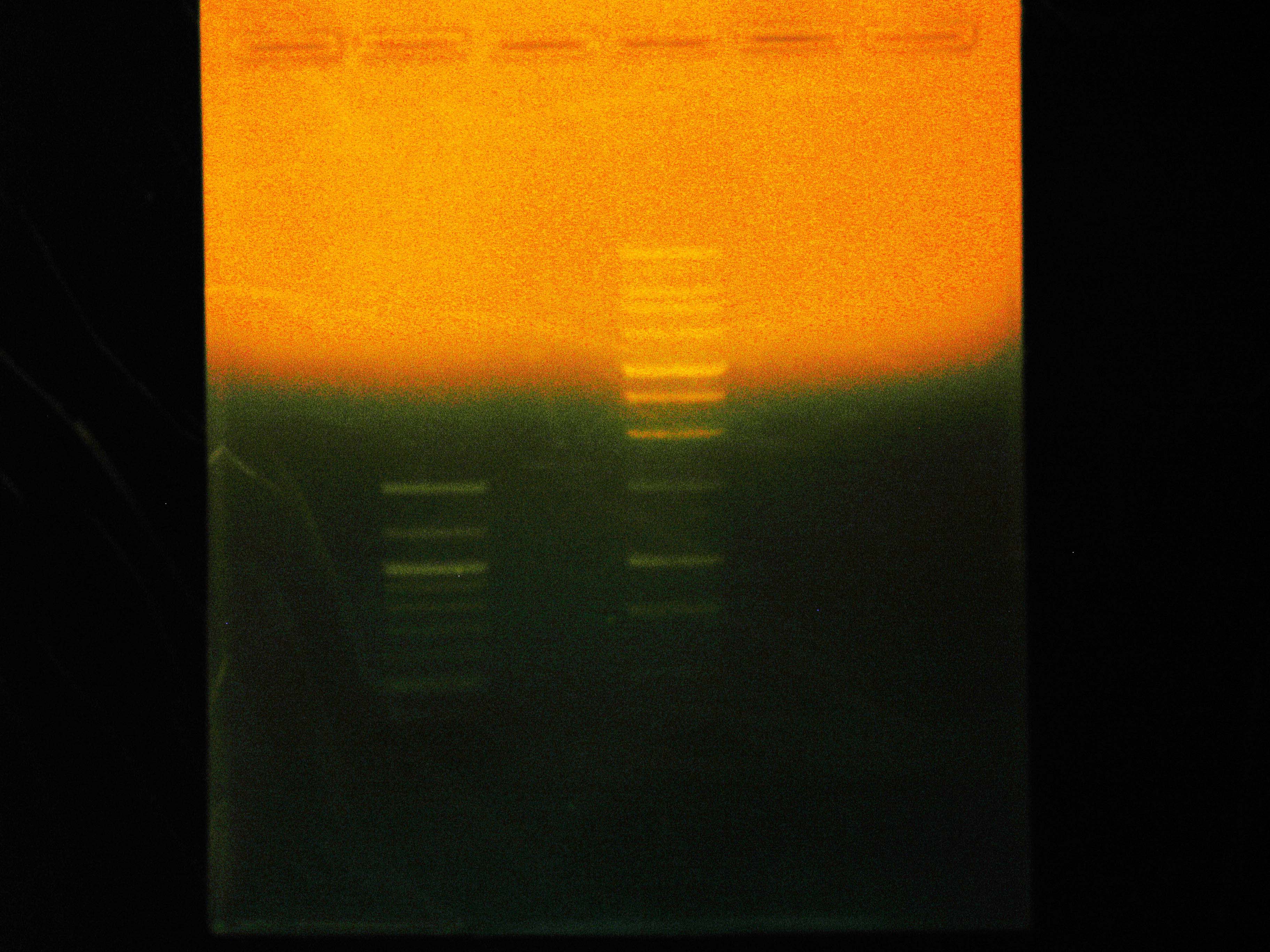
[[File:2012_07_03_EtBrStainGibson.jpg|thumb|Three Gibson Transformants and Three Ladders with Ethidium Bromide Stain After Running the Gel]] | [[File:2012_07_03_EtBrStainGibson.jpg|thumb|Three Gibson Transformants and Three Ladders with Ethidium Bromide Stain After Running the Gel]] | ||
The rerun of our Gibson sequencing failed again, so we tried a more roundabout method of determining whether our 3 Gibson transformants have complete nah operons or not. We digested them each with BamHI, expecting specific fragment lengths on a gel electrophoresis. | The rerun of our Gibson sequencing failed again, so we tried a more roundabout method of determining whether our 3 Gibson transformants have complete nah operons or not. We digested them each with BamHI, expecting specific fragment lengths on a gel electrophoresis. | ||
| Line 49: | Line 47: | ||
We only saw one band on each, all of them at about 3.6kb. | We only saw one band on each, all of them at about 3.6kb. | ||
| - | Because no transformations from our previous competent freezer stocks were successful, Dylan decided to make a starter culture of Shewanella strain JG 700 in preparation for making new competent stocks using the [[PNNL Protocol]]. Swati and Tina used this starter culture to complete the preparation. | + | Because no transformations from our previous competent freezer stocks were successful, Dylan decided to make a starter culture of Shewanella strain JG 700 in preparation for making new competent stocks using the [[Team:Cornell/Notebook/Transformation_shewanella#PNNL|PNNL Protocol]]. Swati and Tina used this starter culture to complete the preparation. |
==July 4th, Wednesday== | ==July 4th, Wednesday== | ||
| - | [[Image:CornellJuly4thBBQ.jpg|thumb|right]] | + | [[Image:CornellJuly4thBBQ.jpg|thumb|right|''"Maneesh Gupta, so delicious"'']] |
In the morning, Dylan, Swati, and Danielle prepared p8, p9, p10, p14, and p15 for sequencing (from the minipreps that Caleb preformed the previous day). | In the morning, Dylan, Swati, and Danielle prepared p8, p9, p10, p14, and p15 for sequencing (from the minipreps that Caleb preformed the previous day). | ||
| - | The team took the rest of the day off and had a barbeque at Buttermilk Falls. There was an excess of food, spontaneous song, and fireflies | + | The team took the rest of the day off and had a barbeque at Buttermilk Falls. There was an excess of food, spontaneous song, and fireflies. |
==July 5th, Thursday== | ==July 5th, Thursday== | ||
| - | Upon arriving to the lab, Dylan dropped off the sequencing tubes we'd set up on the morning of the 4th for analysis. He also decided to do a [[Colony PCR]] of the potential transformant with the Salicylate reporter plasmid. However, when setting up this reaction, Dylan realized that we'd used the wrong sequencing primers (we'd used the standard VF2 BioBrick primer instead of a custom primer for the pBBRBB backbone). Consequently, Mark and Dylan resubmitted p14 and p15 for sequencing while the colony PCR was ongoing. After visualizing the PCR products using [[DNA Gel Electrophoresis]], Dylan concluded that the colony we'd screened did not have our plasmid of interest, and was the result of contamination. | + | Upon arriving to the lab, Dylan dropped off the sequencing tubes we'd set up on the morning of the 4th for analysis. He also decided to do a [[Team:Cornell/Notebook/Colony_PCR|Colony PCR]] of the potential transformant with the Salicylate reporter plasmid. However, when setting up this reaction, Dylan realized that we'd used the wrong sequencing primers (we'd used the standard VF2 BioBrick primer instead of a custom primer for the pBBRBB backbone). Consequently, Mark and Dylan resubmitted p14 and p15 for sequencing while the colony PCR was ongoing. After visualizing the PCR products using [[Team:Cornell/Notebook/Gel_electrophoresis|DNA Gel Electrophoresis]], Dylan concluded that the colony we'd screened did not have our plasmid of interest, and was the result of contamination. |
| - | Similarly, Dylan noticed small, evenly spaced colonies on all of the JG700 transformant plates from the [[Myers and Myers]] procedure we'd undertaken on the 2nd of July. Because none of the colonies looked red (as they should have if they were replicating pSB3C5), Dylan concluded that the chloramphenicol on the plates had degraded, and the colonies were resultant from untransformed cells. | + | [[Image:2012_07_05_ColonyPCR.jpg|left|thumb|Result of the Colony PCR]] |
| + | Similarly, Dylan noticed small, evenly spaced colonies on all of the JG700 transformant plates from the [[Team:Cornell/Notebook/Transformation_shewanella#Myers_and_Myers|Myers and Myers]] procedure we'd undertaken on the 2nd of July. Because none of the colonies looked red (as they should have if they were replicating pSB3C5), Dylan concluded that the chloramphenicol on the plates had degraded, and the colonies were resultant from untransformed cells. | ||
| - | Because the colony PCR of the potential salicylate reporter didn't yield good results, Dylan and Youjin set up another [[Phusion PCR]] to amplify the salicylate-sensing region from p21. Simultaneously, Mark made [[SOB]] media for recovery after future transformations, as we'd just received the ingredients for the broth. | + | Because the colony PCR of the potential salicylate reporter didn't yield good results, Dylan and Youjin set up another [[Team:Cornell/Notebook/Phusion_PCR|Phusion PCR]] to amplify the salicylate-sensing region from p21. Simultaneously, Mark made [[Team:Cornell/Notebook/SOB#Preparing_Super_Optimal_Broth_.28SOB.29|SOB]] media for recovery after future transformations, as we'd just received the ingredients for the broth. |
<!-- | <!-- | ||
What happened to the gel w/ EtBr loaded into the wells???? | What happened to the gel w/ EtBr loaded into the wells???? | ||
--> | --> | ||
| - | + | Because we needed to submit for sequencing twice (because we used the wrong sequencing primers), Swati et al. set up liquid cultures of p21, p8, p9, p10, p17, and p14 in order to have more DNA in the freezer. We like having DNA in the freezer. (See: [[Team:Cornell/Notebook/StrainList|strain list]]) | |
| - | + | ||
| - | Because we needed to submit for sequencing twice (because we used the wrong sequencing primers), Swati et al. set up liquid cultures of p21, p8, p9, p10, p17, and p14 in order to have more DNA in the freezer. We like having DNA in the freezer. | + | |
| - | + | ||
==July 6th, Friday== | ==July 6th, Friday== | ||
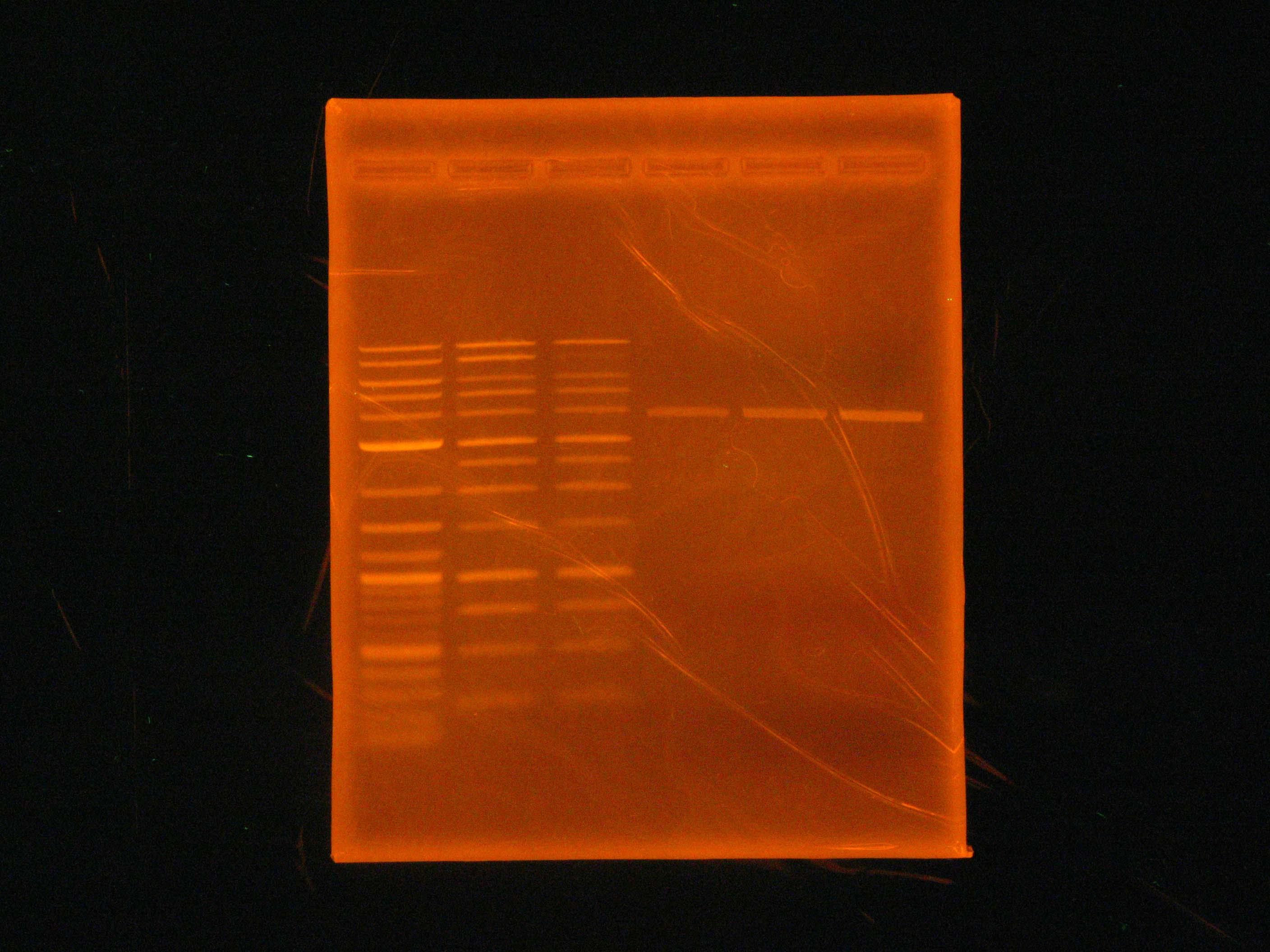
| - | Caleb miniprepped p21, p8, p9, p10, p17 from the liquid cultures set up the previous night. We couldn't miniprep p14, since – after checking notes from the previous night – we learned that the culture was made with the wrong antibiotic (chloramphenicol instead of kanamycin) | + | [[Image:2012_07_06_p21FAIL.jpg|right|thumb|Gel showing the failure of the p21 PCR]] |
| - | + | Caleb [[Team:Cornell/Notebook/Miniprep|miniprepped]] p21, p8, p9, p10, p17 from the liquid cultures set up the previous night. (See: [[Team:Cornell/Notebook/StrainList|strain list]]) We couldn't miniprep p14, since – after checking notes from the previous night – we learned that the culture was made with the wrong antibiotic (chloramphenicol instead of kanamycin). | |
| - | + | ||
| - | + | ||
| + | Some sequencing results came back, however the Gibson results are still confusing. | ||
| - | + | Also, a [[Team:Cornell/Notebook/Gel_electrophoresis|gel]] was run for the p21 PCR product. | |
Latest revision as of 14:02, 16 July 2012
| Home | Team | Project | Parts | Modeling | Notebook | Protocols | Safety | Attributions |
|---|
|
The salicylate reporter is analogous to the arsenic reporter, but with a salicylate-sensitive promoter. That is, in the presence of salicylate it will produce MtrB, completing the Mtr pathway and allowing Shewanella to produce current in our biosensor. In combination with the nah operon, this reporter will be able to detect naphthalene levels. Back to salicylate reporter week view
July 1st-7th
July 1st, Sunday
At 9:50, Dylan and Caleb set up 2 gels for electrophoresis. Caleb's was 1% agarose in BIO-RAD Mini-Sub Cell system for continuation of ladder test using SYBR Green, containing NEB 100 bp ladder, NEB 2-log ladder, Promega 1 kb ladder and run at 100V. Dylan's was 1% agarose in Owl box using ethidium bromide, containing the nah operon PCR product from previous night, and run at 55 V.
Our plates of DH5alpha transformed with p21 ligated into pBBRBB with mtrB grew only one colony, possibly contamination. Dylan ran a colony PCR and got a smear, suggesting that the PCR of p21 or the ligation did not work.
Because we learned that our SYBR Green was causing ladder to run strangely, Dylan decided to redo a Vent PCR to amplify the salicylate reporter region out of p21.
Also, a liquid culture of JG700 was prepared, as well as replating of p21, p22, JG700, JG1220, JG1537, JG1219. (See: strain list)
July 2nd, Monday
Today, Dylan and Caleb ran a gel of a Vent PCR of p21 (the PCR product being the salicylate reporter) at 55 V. Additionally, Caleb decided to run a control gel at 100 V to determine whether higher voltage was a factor in our previous ladder problems (in addition to the SYBR Green stock). While we determined that the higher voltage did not cause our ladder issues, we did not see any bands from the p21 PCR.
Dylan prepared electrocompetent cells using the Myers and Myers protocol. Modified protocol using 2 5mL cultures @ 4000g for 10 min. Washed with 2 mL sorbitol, resuspended in 100 uL sorbitol. First electroporation ts = 2.32 ms, second ts = 2.02 ms. Added 1 uL of plasmid (575.5 ng) to each cuvette. First used .60 V, then 0.55 V, both with R = 400 ohms.
Mark and Danielle started liquid cultures of S1, S9, S10, S11, S15, S16, S18, S27 (See: strain list)
July 3rd, Tuesday
Caleb miniprepped S16 (p15), S22 (p21), S9 (p8), S10 (p9), S11 (p10), S18 (p17), and S15 (p14). (See: strain list) Instead of a single EB elution, Caleb did two elutions, 30 ul each. The double 30 ul elution turned out to be effective at recovering a usable amount of DNA in the second elution, so we're including it on our miniprep protocol for future minipreps.
The rerun of our Gibson sequencing failed again, so we tried a more roundabout method of determining whether our 3 Gibson transformants have complete nah operons or not. We digested them each with BamHI, expecting specific fragment lengths on a gel electrophoresis.
When Dylan and Mark were attempting to determine what volume of ethidium bromide should be used in the gel, they came to the conclusion that better images may result from staining the gel in dilute ethidium bromide after running the gel, rather than including the stain in the gel. This experimental first staining used 200 mL of water conaining 20 uL of 10 mg/mL ethidium bromide, with gentle agitation for 1 hour. This stained the bands well, but also provided a large amount of background and took a very long time.
We only saw one band on each, all of them at about 3.6kb.
Because no transformations from our previous competent freezer stocks were successful, Dylan decided to make a starter culture of Shewanella strain JG 700 in preparation for making new competent stocks using the PNNL Protocol. Swati and Tina used this starter culture to complete the preparation.
July 4th, Wednesday
In the morning, Dylan, Swati, and Danielle prepared p8, p9, p10, p14, and p15 for sequencing (from the minipreps that Caleb preformed the previous day).
The team took the rest of the day off and had a barbeque at Buttermilk Falls. There was an excess of food, spontaneous song, and fireflies.
July 5th, Thursday
Upon arriving to the lab, Dylan dropped off the sequencing tubes we'd set up on the morning of the 4th for analysis. He also decided to do a Colony PCR of the potential transformant with the Salicylate reporter plasmid. However, when setting up this reaction, Dylan realized that we'd used the wrong sequencing primers (we'd used the standard VF2 BioBrick primer instead of a custom primer for the pBBRBB backbone). Consequently, Mark and Dylan resubmitted p14 and p15 for sequencing while the colony PCR was ongoing. After visualizing the PCR products using DNA Gel Electrophoresis, Dylan concluded that the colony we'd screened did not have our plasmid of interest, and was the result of contamination.
Similarly, Dylan noticed small, evenly spaced colonies on all of the JG700 transformant plates from the Myers and Myers procedure we'd undertaken on the 2nd of July. Because none of the colonies looked red (as they should have if they were replicating pSB3C5), Dylan concluded that the chloramphenicol on the plates had degraded, and the colonies were resultant from untransformed cells.
Because the colony PCR of the potential salicylate reporter didn't yield good results, Dylan and Youjin set up another Phusion PCR to amplify the salicylate-sensing region from p21. Simultaneously, Mark made SOB media for recovery after future transformations, as we'd just received the ingredients for the broth.
Because we needed to submit for sequencing twice (because we used the wrong sequencing primers), Swati et al. set up liquid cultures of p21, p8, p9, p10, p17, and p14 in order to have more DNA in the freezer. We like having DNA in the freezer. (See: strain list)
July 6th, Friday
Caleb miniprepped p21, p8, p9, p10, p17 from the liquid cultures set up the previous night. (See: strain list) We couldn't miniprep p14, since – after checking notes from the previous night – we learned that the culture was made with the wrong antibiotic (chloramphenicol instead of kanamycin).
Some sequencing results came back, however the Gibson results are still confusing.
Also, a gel was run for the p21 PCR product.
 "
"