Team:Cambridge/Ribosense/Labbook
From 2012.igem.org
(→Week 7) |
(→Week 14) |
||
| (12 intermediate revisions not shown) | |||
| Line 70: | Line 70: | ||
</html> | </html> | ||
| - | = | + | = Ribosense Labbook = |
==Week 4 == | ==Week 4 == | ||
| Line 427: | Line 427: | ||
==Week 8== | ==Week 8== | ||
| + | |||
| + | ===Wednesday (15/08/12)=== | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/beta-galactosidaseassay| Characterization of fluoride riboswitch construct]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Strain of ''bacillus'' lacking fluoride transporter system tested. Concentrations used: | ||
| + | |||
| + | :*0mM - 0.5mM - 1mM - 2.5mM - 5mM - 10mM - 20mM - 30mM | ||
| + | |||
| + | ===Thursday (16/08/12)=== | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/beta-galactosidaseassay| Characterization of fluoride riboswitch construct]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | [[File:fluoridegradient.jpg|250px|right|thumb|The results of our fluoride assay.]] | ||
| + | |||
| + | * Eppindorfs containing the ''bacillus'' with the fluoride riboswitch removed from incubator and imaged. | ||
| + | |||
| + | * Now this definitely works, we will try quantifying this riboswitch with an ONPG assay and the plate reader. | ||
| + | |||
| + | ===Saturday (18/08/12)=== | ||
| + | |||
| + | '''Colonies picked in preparation for Miller assay''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *2 ''E. coli'' colonies transformed with the purified plasmid were picked and inoculated in LB with 50μg/ml Amp | ||
| + | |||
| + | *2 colonies each from the 168 strain and crcB knockout ''B. subtilis'' containing the fluoride riboswitch were picked and inoculated in LB with 5μg/ml Chlor | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/PCRProtocol|PCR of Fluoride biobrick format]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | [[File:FluorideBB1gel.jpg|right|250px|thumb|Gels from PCR of Fluoride BB and PJS130 fragments. Top: Lanes 2-4: Gibson compatable Fluoride biobrick fragment. Lanes 5-7: Ligation compatable Fluoride biobrick fragment. Lane 8: PJS130 fragment A. Bottom: Lanes 1-2: PJS130 fragment A. Lanes 3-5: PJS 130 fragment B. Lane 6: Positive control. Lane 7: Negative control.]] | ||
| + | |||
| + | *Two sets of primers used: one to allow insertion into backbone by ligation, and one to allow insertion by Gibson assembly. | ||
| + | |||
| + | *Cycle settings: | ||
| + | |||
| + | :*Melting - 98 °C - 10 seconds | ||
| + | |||
| + | :*Annealing - 58 °C - 30 seconds | ||
| + | |||
| + | :*Elongation - 72 °C - 100 seconds | ||
| + | |||
| + | *Products run on gel. Fragments produced of correct size, however they overlapped with the primer dimer band due to the small size of the fragments. | ||
| + | |||
| + | *DNA extracted and purified. | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/PCRProtocol|PCR of Mg2+ riboswitch construct vector]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Separate reactions for | ||
| + | |||
| + | *Cycle settings: | ||
| + | |||
| + | :*Melting - 98 °C - 10 seconds | ||
| + | |||
| + | :*Annealing - 58 °C - 30 seconds | ||
| + | |||
| + | :*Elongation - 72 °C - 100 seconds | ||
| + | |||
| + | *Products run on gel. No fragments produced. Positive control worked however, so master mix clearly works. | ||
| + | |||
| + | *Given this PCR has worked in the past, will try to re-run with the same settings and hope for success in the future. | ||
| + | |||
| + | ===Sunday (19/08/12)=== | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/MillerAssay|Miller Assay of Fluoride construct]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | * Colonies picked from overnight and Miller assay run as directed on protocols page | ||
| + | |||
| + | * Lack of filters meant that only A450 data recorderd | ||
==Week 9== | ==Week 9== | ||
| + | |||
| + | ===Tuesday (21/08/12)=== | ||
| + | |||
| + | '''PCR for Mg Riboswitch''' | ||
| + | |||
| + | ---- | ||
| + | *[[Team:Cambridge/Protocols/PCRProtocol| PCRs]] were run for pJS130.1 split vector ('+8 codons') | ||
| + | *[[Team:Cambridge/Protocols/PCRcolony| Colony PCR]] for endogenous ''Bacillus subtilis'' Mg Riboswitch ('+8 codons') was run | ||
| + | *Gels were run for above reactions, only one band for half of the pJS130 vector was visible. | ||
| + | |||
| + | ===Wednesday (22/08/12)=== | ||
| + | |||
| + | '''PCR for Mg Riboswitch''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *[[Team:Cambridge/Protocols/PCRProtocol| PCRs]] were run for F Riboswitch from Yale's plasmid with overhangs for standard assembly. | ||
| + | *[[Team:Cambridge/Protocols/PCRcolony| Colony PCR]] for endogenous ''Bacillus subtilis'' Mg Riboswitch (both '+8 codons' and '-8 codons' versions) were re-run. | ||
| + | *Gels were run for above reactions, only one band for positive control was visible. | ||
| + | |||
| + | '''Cells cultured for Miller assay''' | ||
| + | ---- | ||
| + | *''B. subtilis'' strains carrying F riboswitch and 'KO' strains cultured separately in LB and 5µg/mL Chloramphenicol at 37°C | ||
| + | *''E.Coli'' carrying F riboswitch cultured in LB and 50mg/ml Ampicillin at 37°C | ||
| + | |||
| + | ===Thursday (23/08/12)=== | ||
| + | * Miller assay set up and running with the wild type and crcB knock-out ''Bacillus'' strains and an ''E. coli'' strain which contains the construct on an episomal plasmid. | ||
| + | |||
| + | ===Friday (24/08/12)=== | ||
| + | *Miller assay data collected for analysis over the weekend | ||
==Week 10== | ==Week 10== | ||
| + | |||
| + | ===Monday (27/08/12)=== | ||
| + | '''RiboSense:[[Team:Cambridge/Protocols/PCRProtocol|PCR of Mg RS from bacillus genomic DNA]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *3 part primers used, to amplify the 486bp from the genome, and the vector in two parts | ||
| + | *PCR failed, only positive control came out. Will re run when new primers arrive. | ||
| + | |||
| + | ===Thursday (30/08/12)=== | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/PCRcolony|PCR of Mg2+ riboswitch from genomic DNA]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | [[File: MGRSgel1.jpg|right|250px|thumb|Gel from amplification of the riboswitch DNA. Lanes 2 + 3: with 8 codon substitution. Lanes 4 + 5: without 8 codon substitution.]] | ||
| + | |||
| + | *Cycle settings: | ||
| + | |||
| + | :*Melting - 98 °C - 10 seconds | ||
| + | |||
| + | :*Annealing - 58 °C - 30 seconds | ||
| + | |||
| + | :*Elongation - 72 °C - 30 seconds | ||
| + | |||
| + | *Fragments of correct size produced for all except lane 5 produced. In this lane, DNA appears to have accumulated in the well, indicating it may be genomic. In future, will use lower numbers of template cells to avoid getting so much genomic DNA. | ||
| + | |||
| + | *Products extracted and purified. | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/PCRProtocol|PCR of PJS130 vector for Mg riboswitch construct]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | [[File:PJS130gel2.jpg|250px|right|thumb|Gel from amplification of PJS130 vector for MGRS construct. Lanes 2 + 3: Fragment A. Lanes 4 + 5: Fragment B with 8 codon substitution. Lanes 6 + 7: Fragment B without 8 codon substitution.]] | ||
| + | |||
| + | *Cycle settings: | ||
| + | |||
| + | :*Melting - 98 °C - 10 seconds | ||
| + | |||
| + | :*Annealing - 60 °C - 30 seconds | ||
| + | |||
| + | :*Elongation - 72 °C - 110 seconds | ||
| + | |||
| + | *Fragment of correct size produced in lane 5, but too faint to be extracted successfully. None of the other lanes were successful. We will try this again tomorrow, at 58 °C and with 35 cycles (as is usual) instead of 30. | ||
| + | |||
| + | *Note that phusion enzyme was left with primers and template DNA for about half an hour before reaction began, possibly causing degradation of the primer DNA. | ||
| + | |||
| + | ===Friday (31/08/12)=== | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/PCRProtocol|PCR of PJS130 vector for MgRS construct]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
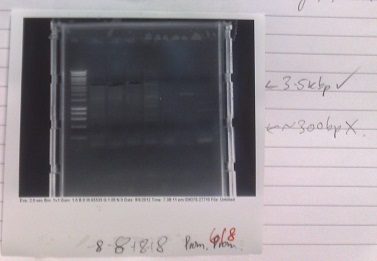
| + | [[File:PJS130gel1.jpg|250px|left|thumb|Gel from amplification of PJS130 fragments. Lanes 2 + 3: Fragment A. Lanes 4 + 5: Fragment B with eight codon substitution. Lanes 6 + 7: Fragment B without eight codon substitution. Lane 8: Negative control.]] | ||
| + | |||
| + | *Cycle settings: | ||
| + | |||
| + | :*Melting - 98 °C - 10 seconds | ||
| + | |||
| + | :*Annealing - 58 °C - 30 seconds | ||
| + | |||
| + | :*Elongation - 72 °C - 120 seconds | ||
| + | |||
| + | *Products of correct sizes (5.5kbp and 3.5 kbp) produced for all reactions, although lanes 2 and 4 failed to produce any product, despite primer smear. Most likely, template was not added, or one of the primers was not added. | ||
| + | |||
| + | *Products excised and purified. | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/Gibsonassembly|Gibson assembly of Mg2+ riboswitch construct]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Reaction 1: Without 8 codon substitution: Vec A, Vec B -8 (replicate 1), Genomic -8. | ||
| + | |||
| + | *Reaction 2: Without 8 codon substitution: Vec A, Vec B -8 (replicate 2), Genomic -8. | ||
| + | |||
| + | *Reaction 3: With 8 codon substitution: Vec A, Vec B +8, Genomic +8 (replicate 1). | ||
| + | |||
| + | *Reaction 4: With 8 codon substitution: Vec A, Vec B +8, Genomic +8 (replicate 2). | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/TransformationofE.coli|Transformation of e.coli with Gibson products]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *20 μl of Gibson reaction mix transformed into e.coli cells. Transformants plated out onto 100 μg/ml ampicillin plates. | ||
| + | |||
| + | ===Saturday (01/09/12)=== | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/GelElectrophoresis|Verification of Mg2+ gel extractions]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | [[File:Mg2+verificationgel1.jpg|250px|right|thumb|Verification gel of extracted DNA. Lanes 1,7 + 13: Ladder. Lanes 2 - 4: Old vector DNA (06/08/12). Lane 5: PJS130 fragment A. Lanes 6,8 + 9: PJS130 fragment B. Lanes 10 - 12: Genomic riboswitch DNA.]] | ||
| + | |||
| + | *DNA used in Gibson from yesterday and several weeks ago. Run on gel to verify the presence of DNA fragments of the correct size after gel extraction. | ||
| + | |||
| + | *Old DNA appears to have degraded, or else products were produced in too small a quantity to show up on the gel. Fortunately, these are not/ have not been used. | ||
| + | |||
| + | *Bands of the correct size for fragment A (5.5kbp), fragment B (3.5kbp) and the genomic DNA (550bp) present. Though riboswitch DNA has not come out well in this image, it was present under visual examination. | ||
| + | |||
| + | *Approximate DNA concentrations in the range of 2 - 6 ng/μl. | ||
| + | |||
| + | ===Sunday (02/09/12)=== | ||
| + | |||
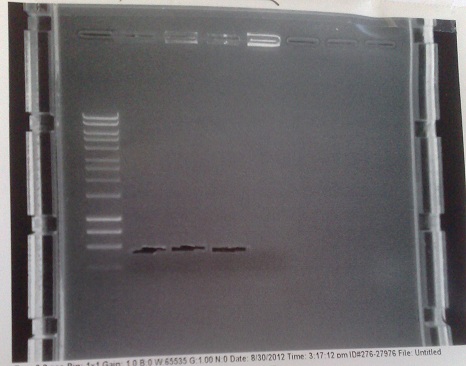
| + | [[File:MGRSverification1.jpg|left|250px|thumb|Results from colony PCR (right of central ladder) and of restriction digest (left of central ladder). Restriction digest produced inconclusive results, however colony PCR indicates that the riboswitch only inserted into the second of the two colonies picked from the +8 version plate (last two lanes before ladder).]] | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/PCRcolony|Verification of Mg2+ riboswitch construct by colony PCR]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Gibson primers used to amplify out riboswitch section of plasmid produced and transformed into e.coli to verify if riboswitch had inserted into PJS130. | ||
| + | *Band at ~550bp in lanes 8 and 9 (replicates) indicate that the riboswich has only been successfuly transformed into the second of the two colonies picked on the +8 plate, and not at all for the -8 construct. | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/MiniPrep|Miniprep of plasmid DNA]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *E.coli from strains made yesterday miniprepped to get plasmid DNA at high concentration. After colony PCR results, all DNA other than that of the successful colony discarded. | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/RestrictionDigest|Verification of Mg2+ riboswitch construct by restriction digest]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *BamH1 and Sal1 used to digest plasmid DNA produced from miniprep. | ||
| + | *Resultant fragment run on gel. | ||
| + | *Bands produced inconclusive, possibly because of too short an incubation time with restriction enzymes. Expected bands at ~4900bp and ~4500bp. Will re-run experiment in future to check results of colony PCR. | ||
==Week 11== | ==Week 11== | ||
| + | |||
| + | ===Wednesday (05/09/12)=== | ||
| + | |||
| + | [[File:BBPCR1.jpg|250px|left|thumb|PCR products from Wednesday's PCR. Lanes 2 - 3: Fluoride riboswitch biobrick genomic DNA. Lanes 4 - 5: MGRS construct genomic DNA (+8). Lanes 6 - 7: MGRS construct genomic DNA (-8). Lane 8: MGRS biobrick genomic DNA (+8).]] | ||
| + | |||
| + | [[File:BBPCR2.jpg|250px|right|thumb|PCR products from Wednesday's PCR. Top: Lane 2: MGRS biobrick genomic DNA (+8). Lanes 3 - 4: MGRS construct backbone (expected size 9kbp) (+8). Lanes 5 - 6: MGRS construct backbone (expected size 9kbp) (-8). Lane 7: MGRS biobrick vector (+8). Lane 8: Positive control. Bottom: Lane 2: MGRS biobrick vector (+8). lane 3: Negative control.]] | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/PCRProtocol|PCR of biobrick fragments and MGRS construct fragments.]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Fluoride and magnesium riboswitch genomic DNA amplified with PCR, along with vector fragments for turning the MGRS into a biobrick and for producing the magnesium riboswitch construct. | ||
| + | *PCR settings: Annealing temperature - 58 °C, Elongation step - 60 seconds. | ||
| + | *Genomic fragments worked well, but longer backbone fragments mostly failed. | ||
| + | *Extractions of DNA from these gels failed almost completely. Will try purifying directly from PCR products next time - results may not produce such a high purity of the precise DNA we want, but the increase in yield should outweigh this problem. | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/Gibsonassembly|Gibson assembly of -8 MGRS construct]] | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Fragments gel extracted earlier assembled with Gibson assembly. | ||
| + | *Fragments used: | ||
| + | :*9kbp fragment from tube 13 used as backbone. | ||
| + | :*Genomic fragments from tubes 6 + 7. | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/TransformationofE.coli|Transformation of E.coli with Gibson products]] | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Gibson products from earlier today used to transform e.coli. | ||
| + | *Resultant transformants plated out on 100 μg/ml ampicillin plates. | ||
==Week 12== | ==Week 12== | ||
| + | |||
| + | ===Monday (10/09/12)=== | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/PCRProtocol|PCR of biobrick vector DNA]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Attempted to run PCR of the backbone for the biobricks once more. Settings: Annealing temperature: 56°C, elongation time: 30secs. | ||
| + | |||
| + | *After running on gel, saw that while the -8 vector amplification appears to be producing some correctly sized (~2kb) bands, the other two only seem to be forming many primer dimers. Analysis of the sequence of the prefix and suffix revealed a CG palindrome that appears to be causing self annealing. Inserts are not affected, as they have the palindrome at the 5' end of the primer. | ||
| + | |||
| + | *Will retry at higher temperatures tomorrow. | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/Gibsonassembly|Gibson assembly of biobricks]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Attempted Gibson assembly for the -8 magnesium riboswitch biobrick with the backbone fragments produced earlier today and the insert fragments produced yesterday. | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/TransformationofE.coli|Transformation of e.coli with Gibson products]] | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *E.coli cells transformed with Gibson products produced earlier today. | ||
| + | |||
| + | *Transformants plated out on 25μ/ml chloramphenicol plates. | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/RestrictionDigest|Restriction digest of magnesium riboswitch construct plasmids]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Colonies grown up from colonies produced from Gibson products made on '''Check!!!''' were miniprepped and plasmid DNA extracted. | ||
| + | |||
| + | *Restriction digest performed with Sal1 and BamH1. Expected fragment sizes: 4900bp and 4500bp. If plasmid lacks insert: 4900bp and 3950bp. These two possibilities should be distinguishable. | ||
| + | |||
| + | *Correct banding pattern seen for all colonies grown up, with both -8 and +8 construct. Shall verify identity further with colony PCR. | ||
| + | |||
| + | ===Thursday (13/09/12)=== | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/MgFreeCells#Production_of_Magnesium_Free_Cells|Magnesium Riboswitch Plate-Reader Assay]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | *Plate reader assay set up with magnesium riboswitch construct made over the last few days. | ||
| + | |||
| + | *Mg2+ concentrations used: | ||
| + | |||
| + | :*5μM, 10μM, 20μM, 50μM, 100μM, 200μM, 500μM, 1mM, 2mM, 5mM, 10mM, 20mM | ||
| + | |||
| + | *IPTG concentrations used: | ||
| + | |||
| + | :*0.05mM, 0.1mM, 0.2mM, 0.5mM, 1mM, 2mM, 5mM, 10mM | ||
| + | |||
| + | *Magnesium free medium B used as growth medium | ||
| + | |||
| + | *Results: fluorescence curves are constant, except for a distinct dip during the exponential phase of bacterial growth. After looking at various cultures under the fluorescence microscope and discussing the results with our advisors, we have decided that this is because the amino acids in the medium are fluorescing. Our bacteria are then degrading these, causing the dip observed. We will switch to M9 minimal medium for our assays in the future, as this does not autofluoresce. | ||
==Week 13== | ==Week 13== | ||
==Week 14== | ==Week 14== | ||
| + | |||
| + | ===Wednesday (26/09/12)=== | ||
| + | |||
| + | '''[[Team:Cambridge/Protocols/MillerAssay|Miller Assay of Fluoride construct]]''' | ||
| + | |||
| + | ---- | ||
| + | |||
| + | * colonies picked from overnight and Miller assay run again as directed on protocols page | ||
| + | |||
| + | * 2 colonies each of ''E. coli'', 168 strain, and crcB knockout strain ''Bacillus'' assayed | ||
| + | |||
| + | * Greater resolution at lower fluoride concentrations to test theory from previous assay | ||
| + | |||
| + | * Lack of filters meant that only A450 data recorderd | ||
<html> | <html> | ||
Latest revision as of 03:34, 27 October 2012
Contents |
Judging Form
- Please help the judges by filling out this form. Tell them what medal you think you deserve and why. Tell them which special prizes you should win. Help them find your best parts. Show them how you thought about the safety of your project. Helping the judges will help you too.
- Team: Cambridge
- Region: Europe
- iGEM Year:2012
- Track:Foundational Advance
- Project Name:Parts for a reliable and field ready biosensing platform
- Project Abstract: Implementation of biosensors in real world situations has been made difficult by the unpredictable and non-quantified outputs of existing solutions, as well as a lack of appropriate storage, distribution and utilization systems. This leaves a large gap between a simple, functional sensing mechanism and a fully realised product that can be used in the field.
We aim to bridge this gap at all points by developing a standardised ratiometric luciferase output in a Bacillus chassis. This output can be linked up with prototyped instrumentation and software for obtaining reliable quantified results. Additionally, we have reduced the specialized requirements for the storage and distribution of our bacteria by using Bacillus' sporulation system. To improve the performance of our biosensing platform we have genetically modified Bacillus’ germination speed. Lastly, we demonstrated the robustness of our system by testing it with a new fluoride riboswitch, providing the opportunity to tackle real life problems.
iGEM Medals for non-software teams
- We believe our team deserves the following medal:
- Bronze
- Silver
- √Gold
Because we met the following criteria (check all that apply and provide details where needed)
Requirements for a Bronze Medal
- √Register the team, have a great summer, and plan to have fun at the Regional Jamboree.
- √Successfully complete and submit this iGEM 2012 Judging form.
- √Create and share a Description of the team's project using the iGEM wiki and the team's parts using the Registry of Standard Biological Parts.
- √Plan to present a Poster and Talk at the iGEM Jamboree.
- √Enter information detailing at least one new standard BioBrick Part or Device in the Registry of Standard Biological Parts. Including:
- √Primary nucleaic acid sequence
- √Description of function
- √Authorship
- Safety notes, if relevant.
- √Acknowedgment of sources and references
- √Submit DNA for at least one new BioBrick Part or Device to the Registry.
Additional Requirements for a Silver Medal
- √Demonstrate that at least one new BioBrick Part or Device of your own design and construction works as expected; characterize the operation of your new part/device.
- √Enter this information and other documentation on the part's 'Main Page' section of the Registry
Part Number(s): [http://partsregistry.org/Part:BBa_K911004 BBa_K911004]
Additional Requirements for a Gold Medal: (one OR more)
- Improve an existing BioBrick Part or Device and enter this information back on the Experience Page of the Registry.
Part Number(s): None - √Help another iGEM team by, for example, characterizing a part, debugging a construct, or modeling or simulating their system.
Link to this information on your wiki. Page name: Team:Cambridge/Outreach/Collaboration - √Outline and detail a new approach to an issue of Human Practice in synthetic biology as it relates to your project, such as safety, security, ethics, or ownership, sharing, and innovation.
Link to this information on your wiki.
Page name: Team:Cambridge/HumanPractices/Overview,Team:Cambridge/HumanPractices/MarketResearch,Team:Cambridge/HumanPractices/FutureDirections
iGEM Prizes
All teams are eligible for special prizes at the Jamborees. more... To help the judges, please indicate if you feel you should be evaluated for any of the following special prizes:
- √Best Human Practice Advance
- √Best Experimental Measurement
- Best Model
Please explain briefly why you should receive any of these special prizes:
Best Human Practice Advance:
We feel that we deserve this prize for three reasons:
- We explored the impacts, *both positive and negative*, of synthetic biology as a solution to real world problems, through interviewing professionals working in a relevant field, namely the impact of arsenic water contamination in Bangladesh.
- We recognized existing problems with the way the current direction of synthetic. On going through the registry we found that most of the characterization data for biosensing parts is often neither comparable nor replicable. We have worked to solve this issue, for example with our ratiometric dual channel output.
- *Our project doesn’t stop here*, in Chanel number 6 (Team:Cambridge/HumanPractices/FutureDirections) we considered the future implications and technological applications of our project, as well as the means by which it could be improved by subsequent users. We feel that the end to an iGEM project should not be the conclusion of an idea, but the start of it.
Best BioBrick Measurement Approach:
It is absolutely vital that a quantitative, numerical, robust, and flexible measurement approach exists to relay information to a user that is an accurate representation of the input processed by a biological device. Working from these principles, the following was done:
- We designed and built Biologger, a *cheap, arduino-based, fully functional automatic rotary device* that has an incorporated ratiolumnometer
- Our project is entirely open-sourced and open-platform. We have published source code for the two applications which serve to operate the device, one for PCs and the other for Android devices, as well as the open source circuit design that provides this ratiometric reading. Furthermore, the Android app is able to receive its data wirelessly, which we feel is a great advance in BioBrick measurement.
- Our dual-channel luciferase reporter was successfully tested with a dilution series of E.coli transformed with the Lux Operon (under pBAD) biobrick (Part BBa_K325909) of the Cambridge iGEM 2010 team. It can detect, with good accuracy, both different light intensities, as well as the percentages of blue or orange frequencies in a sample.
- Our device was successfully tested using artificial light to detect different frequencies (colours) as well.
Having done all the above, we believe that this fully open-sourced instrumentation kit (mechanical) chassis, electronics, software code), estimated at *$35.00* (or $85.00 if a Bluetooth modem is required), is a complete BioBrick measurement solution for any and all BioBricks with a light output.
Team_Parts
To help the judges evaluate your parts, please identify 3 of your parts that you feel are best documented and are of the highest quality.
- Best new BioBrick part (natural)
- [http://partsregistry.org/Part:BBa_K911003 BBa_K911003]
- Best new BioBrick part (engineered)
- [http://partsregistry.org/Part:BBa_K911004 BBa_K911004]
- Best improved part(s): None
List any other parts you would like the judges to examine:[http://partsregistry.org/Part:BBa_K911001 BBa_K911001], [http://partsregistry.org/Part:BBa_K911008 BBa_K911009], [http://partsregistry.org/Part:BBa_K911008 BBa_K911008]
Please explain briefly why the judges should examine these other parts:
- Magnesium Sensitive Riboswitch [http://partsregistry.org/Part:BBa_K911001 BBa_K911001]
As a riboswitch sensing construct, this part is an entirely new type of biosensor (along with the fluoride construct) that could potentially change the way we think about designing input genetic circuits. Unlike the fluoride riboswitch, it is a derepression system and therefore serves to demonstrate the principle that riboswitches can be used regardless of whether they turn on or off their reporter. - Fluorescent ratiometric construct for standardizing promoter output [http://partsregistry.org/Part:BBa_K911009 BBa_K911009]
Fluorescence is a major cornerstone for biosensors in the registry, however, most parts do not involve the use of a ratiometric output, which has been shown in the literature to provide much more reliable and meaningful data. This part not only furthers the development of ratiometric measurements in molecular biology but due to the choice of promoters and terminators it can be used to characterize the difference in activity between E. coli and B. Subtilis - Fast Germination (B. subtilis) [http://partsregistry.org/Part:BBa_K911008 BBa_K911008]
This part is entirely novel for the registry and fully utilizes the recombination machinery inherent in the Bacillus chassis. Have spores that can germinate at a faster rate is certainly a worthy achievement and could help with experiments with B. Subtilis that any future iGEM teams may wish to perform.
iGEM Safety
For iGEM 2012 teams are asked to detail how they approached any issues of biological safety associated with their projects.
The iGEM judges expect that you have answered the four safety questions Safety page on your iGEM 2012 wiki.
Please provide the link to that page: Page name: Team:Cambridge/Safety
Attribution and Contributions
For iGEM 2012 the description of each project must clearly attribute work done by the team and distinguish it from work done by others, including the host labs, advisors, and instructors.
Please provide the link to that page, or comments in the box below: Page name: Team:Cambridge/Attributions
Comments
If there is any other information about your project you would like to highlight for the judges, please provide a link to your wiki page here: Team:Cambridge/Overview/DesignProcess
Ribosense Labbook
Week 4
Tuesday (17/07/12)
- Plasmid PJS 130 used as our backbone for isolating the magnesium riboswitch.
- Primers:
- Forward: TTCAAAACATGACCTATGACgtcgcagagtatgccg
- Reverse: cctccctctgctaaaacACAAGGCATTAACACTACAT
- PCR settings:
- 95 °C - 6mins
- 98 °C - 10secs
- 58 °C - 45secs
- 72 °C - 180secs
- Repeat above 35x
- 72 °C - 5mins
- 25 °C - 1min
PCR of riboswitch DNA from bacillus genome
- Colony of strain 168 used as template for this experiment.
- Primers used:
- Forward: cggagggagacgattttgTGTTCCGTAATTGTGATGTAAG
- Reverse: acaccggcatactctgcgacGTCATAGGTCATGTTTTGAACC
- PCR settings - as above (run in parallel).
Wednesday (18/07/12)
Gel electrophoresis of PCR products
- PCR fragments for Mg2+ riboswitch from yesterday separated on gel. 90mins at 100V.
- Lane 1: Ladder
- Lane 2 + 3: Riboswitch DNA from genome (replicates) - 550bp fragment expected.
- Lane 4 + 5: Vector DNA from pJS130 (replicates)- Chloramphenicol resistance marker - 9.0kbp fragment expected
- Lane 6: Postitive control - 1.9kbp fragment expected
- Riboswitch DNA was successfully amplified. However, vector DNA was not. Given that the postive control was also successful, it seems likely that something went wrong with some stage before the PCR amplification. We will try to rectify the problem tomorrow.
Purification of riboswitch DNA from gel
- Successful riboswitch DNA extracted from gel and purified using a minelute column. DNA frozen for later Gibson assembly.
Thursday (19/07/12)
- Plasmid PJS 130 used as our backbone for isolating the magnesium riboswitch.
- Primers:
- Forward: TTCAAAACATGACCTATGACgtcgcagagtatgccg
- Reverse: cctccctctgctaaaacACAAGGCATTAACACTACAT
- PCR settings:
- 95 °C - 6mins
- 98 °C - 10secs
- 55 °C - 45secs
- 72 °C - 240secs
- Repeat above 35x
- 72 °C - 5mins
- 25 °C - 1min
- Settings changed - Annealing temperature and elongation time - attempting to debug PCR.
Gel electrophoresis of PCR products
- PCR fragments for Mg2+ riboswitch from yesterday separated on gel. 90mins at 100V.
- Lane 1: Ladder
- Lane 2, 3 + 4: Vector DNA from pJS130 (replicates) - 9.0kbp fragment expected
- Lane 5: Postitive control - 1.9kbp fragment expected
- Lane 6: Negative control - no fragments expected
- Once more, the PCR appears to have failed. We will try to redesign the primers tomorrow to make them functional.
Week 5
Thursday (26/07/12)
PCR of riboswitch DNA from bacillus genome and Lux genes from e.coli genome
- Colony of strain pJS130 used as template for riboswitch, colony of previously transformed e.coli used as template for lux operon.
- M2+ RS
- Forward: cggagggagacgattttg|TGTTCCGTAATTGTGATGTAAG
- Reverse: tataacgttactggtttcat|CGGGACTCGTACCTCC
- Lux operon
- Forward: gctgtacaag|GGAGGAGGAGGAAGTGGAGGAGGAGGAAGT|atgaagtttggaaatatttgtttttc
- Reverse: cctcgcccttgctcaccat|ACTCTATTCCTTTTTGGTGATTC
- PCR settings - as above (run in parallel).
Friday (27/07/12)
Gel electrophoresis of PCR products
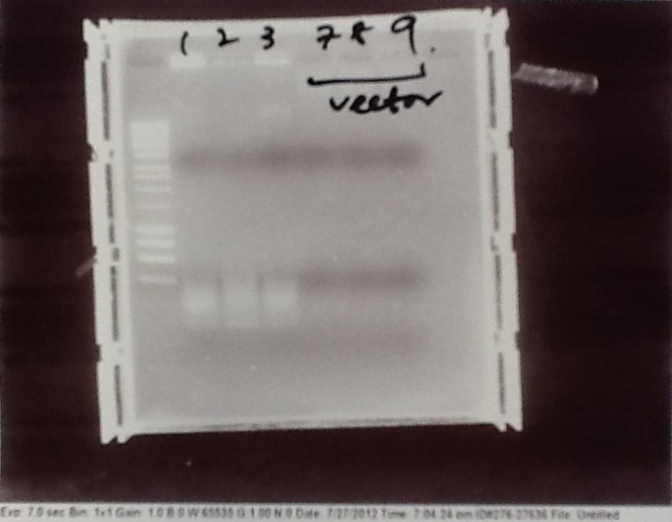
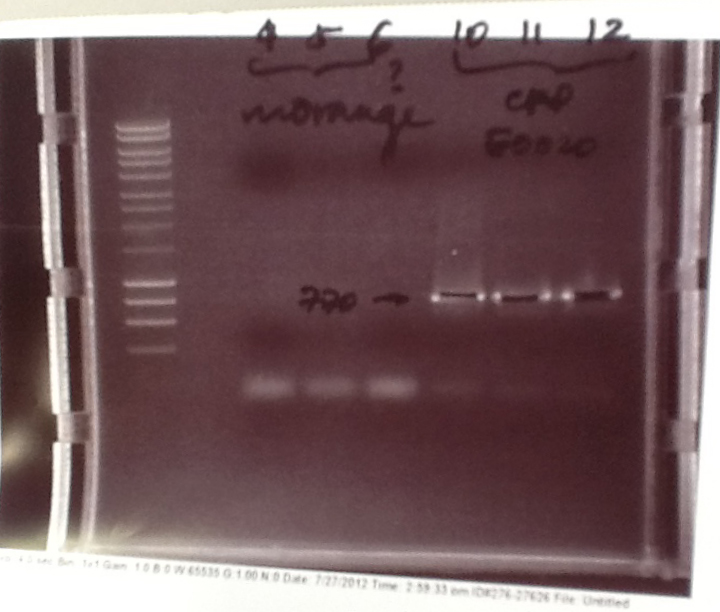
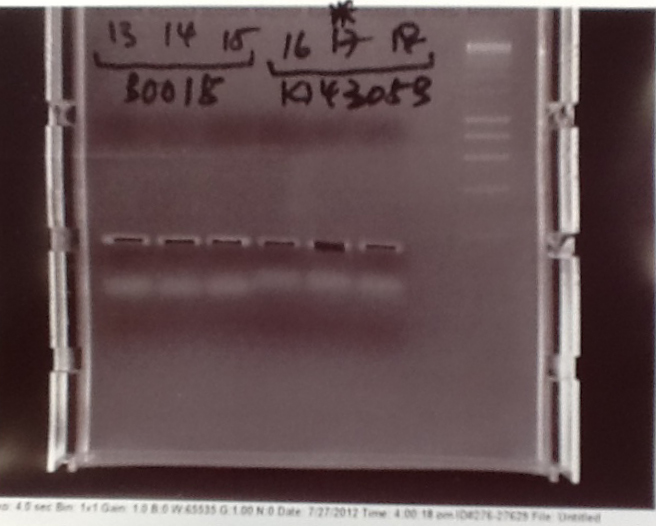
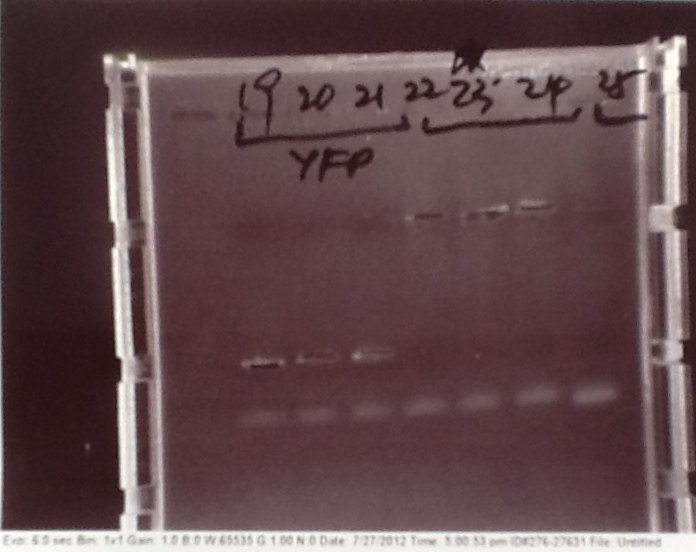
- PCR fragments for Mg2+ riboswitch, luxA/mOrange fusion and fluorescent construct from yesterday separated on gels. 90mins at 100V.
- Mostly successful, but will need to repeat four runs: Three vectors (fluorescent, fusion (lux) and Mg2+ riboswitch (-8 codons) and mOrange gene.
Purification of successfully amplified DNA from gel
- Successful CFP, YFP, B0015, Pveg + SpoVG (K143053), riboswitch and vector DNA extracted from gel and purified using a minelute column. DNA frozen for later Gibson assembly.
Week 6
Monday (30/07/12)
- Not all products were obtained during Friday's PCR. Most of these missing products were large vector backbones. They are being run again, with a much longer extension time of 300s. If that fails, primers will be ordered to split the vectors into manageable chunks, and the PCR reattempted when they arrive.
- PCR cycle x35:
- 15s Denaturing at 95 C
- 45s Annealing at 60 C
- 300s Extension at 72 C
- Remaining stray product had a slightly tricky secondary structure at the 3' end. It will be run at a series of annealing temperatures in a PCR machine capable of a temperature gradient.
- mOrange PCR run at many different temperatures, from 62 °C to 76 °C. However this doesn't seem to solve our problem (refer to Gel photos). It seems likely that it is a primer design problem.
Construction of riboswitch plasmid with Gibson Assembly
- DNA from lanes 27+28, 27+29, 27+30 from gels run on Friday fused together with Gibson assembly to produce riboswitch construct. This does not have replacement of the first 8 codons of lac I with the 8 codons native to the gene downstream of the riboswitch.
- DNA from lanes 22,23 and 24 fused with riboswitch DNA produced two weeks ago to produce riboswitch construct. This has replacement of the first 8 codons of lac I with the 8 codons native to the gene downstream of the riboswitch.
Transformation of Bacillus with riboswitch construct
- Plasmids made by Gibson transformed into bacillus cells made two weeks ago and transformants plated out on 5μg/ml chloramphenicol plates.
Tuesday (01/08/12)
Transformation of Bacillus with riboswitch construct
- Plasmids made by Gibson transformed into bacillus cells made two weeks ago and transformants plated out on 5μg/ml chloramphenicol plates.
Transformation of TOP10 E.coli with riboswitch construct
- Plasmids made by Gibson transformed into TOP10 e.coli cells and transformants plated out on 100μg/ml ampicillin plates.
Wednesday (01/08/12)
Mg2+ Riboswitch
- Successful colonies produced from transformations two days ago streaked out onto chloramphenicol (5 μg/ml) containing plates.
- Colonies also grown up in 10ml of medium A for use with plate reader later.
===Friday (03/08/12)
- PCR of magnesium riboswitch vector repeated with primers to split plasmid.
- Lanes 2 + 3: Fragment A (center - cut site (promotor side))
- Lanes 4 + 5: Fragment B (without 8 codon substitution) (cut site (lac I side) - center)
- Lanes 6 + 7: Fragment B (with 8 codon substitution) (cut stie (lac I side) - center)
- Gels run, found PCR was unsuccessful.
Sunday (05/08/12)
Ribosense: PCR of Mg Riboswitch vector fragments
- 2nd attempt since the PCR on Friday did not work, possibly due to mastermix problems
- PCR Programme:
- Results: Fragment A was successfully PCR-ed (lanes 3-4); Fragment B did not come out, with or without the 8 codons (lanes 5-6 (-8); lanes 7-8 (+8))
Week 7
Monday (06/08/12)
PCR of Magnesium riboswitch vector fragment B and Magnesium promoter
- Normal PCR settings used, annealing temperature 57 °C, elongation step 90s long.
- Lane 5 accidentally loaded with a DNA ladder instead of loading dye.
- Expected fragment sizes:
- Lane 2-5: 3kbp
- Lane 6-7: 300bp
- After electrophoresis, found vector products had, for the most part, worked. Promoter elements were not amplified - no band of the expected size was observed.
- Positive control also failed, although this has ceased to work for several days, potentially due to DNA degradation.
Tuesday (07/08/12)
Gibson assembly of magnesium riboswitch
- NAD+ added to isothermal buffer*5 mix
- Gel slices from yesterday (of vector fragment B) purified.
- DNA added as follows:
- Without 8 codon substitution:
- Reaction 1: Tube 2 (07/08/12) (fragment B), Tube 1 (05/08/12) (fragment A), Tube 28 (29/08/12) (riboswitch DNA).
- Reaction 2: Tube 3 (07/08/12) (fragment B), Tube 2 (05/08/12) (fragment A), Tube 29 (29/08/12) (riboswitch DNA).
- With 8 codon substitution:
- Reaction 3: Tube 4 (07/08/12) (fragment B), Tube 1 (05/08/12) (fragment A), Tube 2 (18/07/12) (riboswitch DNA).
Wednesday (08/08/12)
Electrical transformation of competent e.coli with Gibson products
- Gibson constructs from yesterday (fluorescent and magnesium riboswitch) transformed into e.coli produced yesterday.
- Cells plated out onto 50 μg/ml ampicillin plates. Put in incubator overnight.
Friday (10/08/12)
Characterization of fluoride riboswitch sensitivity with β-galactosidase
- X-Gal made up to a concentration of 400 μg/ml with water.
- Fluoride of concentrations 0.5 - 30 mM mixed with bacterial culture (Yale construct containing) grown up overnight.
Saturday (11/08/12)
Restriction digest of Yale plasmid
- Cells from electroporation of e.coli with plasmid from Yale (08/08/12) miniprepped to extract plasmid DNA.
- DNA digested with XhoI and SalI, expected fragment sizes 5580, 3139, 1437
- Gel run, results not shown as gel was very 'messy'. Gel had probably already begun to set when poured and so had many artefacts when imaged. As a result each lane required different brightness and contrast settings to visualise and so the full gel was not clearly visible on the printout. However, all of the bands could be identified on the screen and were exactly as expected and very clear and pure.
- Bands in correct places, indicating that expected plasmid was present in the cells. This indicates that, although they work at a very low efficiency, our electro-competent cells still work. It may be valuable to use these when trying to grow up plasmids that we already have at high concentration.
Transformation of e.coli with Magnesium riboswitch construct
- Gibson products from 07/08/12 transformed into chemically competent cells.
- Transformants plated out on 100 μg/ml ampicillin plates
Week 8
Wednesday (15/08/12)
Characterization of fluoride riboswitch construct
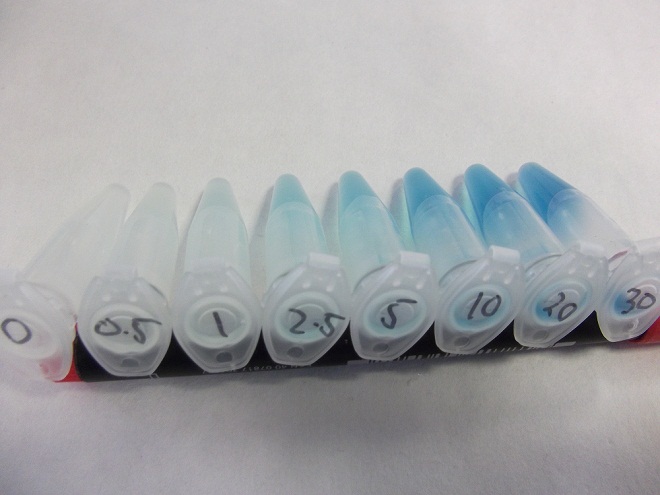
- Strain of bacillus lacking fluoride transporter system tested. Concentrations used:
- 0mM - 0.5mM - 1mM - 2.5mM - 5mM - 10mM - 20mM - 30mM
Thursday (16/08/12)
Characterization of fluoride riboswitch construct
- Eppindorfs containing the bacillus with the fluoride riboswitch removed from incubator and imaged.
- Now this definitely works, we will try quantifying this riboswitch with an ONPG assay and the plate reader.
Saturday (18/08/12)
Colonies picked in preparation for Miller assay
- 2 E. coli colonies transformed with the purified plasmid were picked and inoculated in LB with 50μg/ml Amp
- 2 colonies each from the 168 strain and crcB knockout B. subtilis containing the fluoride riboswitch were picked and inoculated in LB with 5μg/ml Chlor
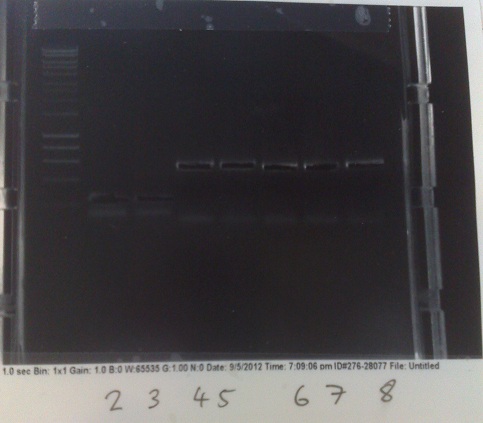
PCR of Fluoride biobrick format
- Two sets of primers used: one to allow insertion into backbone by ligation, and one to allow insertion by Gibson assembly.
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 58 °C - 30 seconds
- Elongation - 72 °C - 100 seconds
- Products run on gel. Fragments produced of correct size, however they overlapped with the primer dimer band due to the small size of the fragments.
- DNA extracted and purified.
PCR of Mg2+ riboswitch construct vector
- Separate reactions for
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 58 °C - 30 seconds
- Elongation - 72 °C - 100 seconds
- Products run on gel. No fragments produced. Positive control worked however, so master mix clearly works.
- Given this PCR has worked in the past, will try to re-run with the same settings and hope for success in the future.
Sunday (19/08/12)
Miller Assay of Fluoride construct
- Colonies picked from overnight and Miller assay run as directed on protocols page
- Lack of filters meant that only A450 data recorderd
Week 9
Tuesday (21/08/12)
PCR for Mg Riboswitch
- PCRs were run for pJS130.1 split vector ('+8 codons')
- Colony PCR for endogenous Bacillus subtilis Mg Riboswitch ('+8 codons') was run
- Gels were run for above reactions, only one band for half of the pJS130 vector was visible.
Wednesday (22/08/12)
PCR for Mg Riboswitch
- PCRs were run for F Riboswitch from Yale's plasmid with overhangs for standard assembly.
- Colony PCR for endogenous Bacillus subtilis Mg Riboswitch (both '+8 codons' and '-8 codons' versions) were re-run.
- Gels were run for above reactions, only one band for positive control was visible.
Cells cultured for Miller assay
- B. subtilis strains carrying F riboswitch and 'KO' strains cultured separately in LB and 5µg/mL Chloramphenicol at 37°C
- E.Coli carrying F riboswitch cultured in LB and 50mg/ml Ampicillin at 37°C
Thursday (23/08/12)
- Miller assay set up and running with the wild type and crcB knock-out Bacillus strains and an E. coli strain which contains the construct on an episomal plasmid.
Friday (24/08/12)
- Miller assay data collected for analysis over the weekend
Week 10
Monday (27/08/12)
RiboSense:PCR of Mg RS from bacillus genomic DNA
- 3 part primers used, to amplify the 486bp from the genome, and the vector in two parts
- PCR failed, only positive control came out. Will re run when new primers arrive.
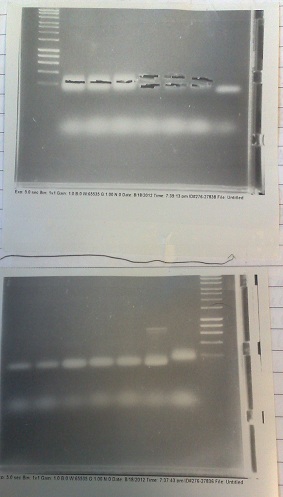
Thursday (30/08/12)
PCR of Mg2+ riboswitch from genomic DNA
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 58 °C - 30 seconds
- Elongation - 72 °C - 30 seconds
- Fragments of correct size produced for all except lane 5 produced. In this lane, DNA appears to have accumulated in the well, indicating it may be genomic. In future, will use lower numbers of template cells to avoid getting so much genomic DNA.
- Products extracted and purified.
PCR of PJS130 vector for Mg riboswitch construct
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 60 °C - 30 seconds
- Elongation - 72 °C - 110 seconds
- Fragment of correct size produced in lane 5, but too faint to be extracted successfully. None of the other lanes were successful. We will try this again tomorrow, at 58 °C and with 35 cycles (as is usual) instead of 30.
- Note that phusion enzyme was left with primers and template DNA for about half an hour before reaction began, possibly causing degradation of the primer DNA.
Friday (31/08/12)
PCR of PJS130 vector for MgRS construct
- Cycle settings:
- Melting - 98 °C - 10 seconds
- Annealing - 58 °C - 30 seconds
- Elongation - 72 °C - 120 seconds
- Products of correct sizes (5.5kbp and 3.5 kbp) produced for all reactions, although lanes 2 and 4 failed to produce any product, despite primer smear. Most likely, template was not added, or one of the primers was not added.
- Products excised and purified.
Gibson assembly of Mg2+ riboswitch construct
- Reaction 1: Without 8 codon substitution: Vec A, Vec B -8 (replicate 1), Genomic -8.
- Reaction 2: Without 8 codon substitution: Vec A, Vec B -8 (replicate 2), Genomic -8.
- Reaction 3: With 8 codon substitution: Vec A, Vec B +8, Genomic +8 (replicate 1).
- Reaction 4: With 8 codon substitution: Vec A, Vec B +8, Genomic +8 (replicate 2).
Transformation of e.coli with Gibson products
- 20 μl of Gibson reaction mix transformed into e.coli cells. Transformants plated out onto 100 μg/ml ampicillin plates.
Saturday (01/09/12)
Verification of Mg2+ gel extractions
- DNA used in Gibson from yesterday and several weeks ago. Run on gel to verify the presence of DNA fragments of the correct size after gel extraction.
- Old DNA appears to have degraded, or else products were produced in too small a quantity to show up on the gel. Fortunately, these are not/ have not been used.
- Bands of the correct size for fragment A (5.5kbp), fragment B (3.5kbp) and the genomic DNA (550bp) present. Though riboswitch DNA has not come out well in this image, it was present under visual examination.
- Approximate DNA concentrations in the range of 2 - 6 ng/μl.
Sunday (02/09/12)
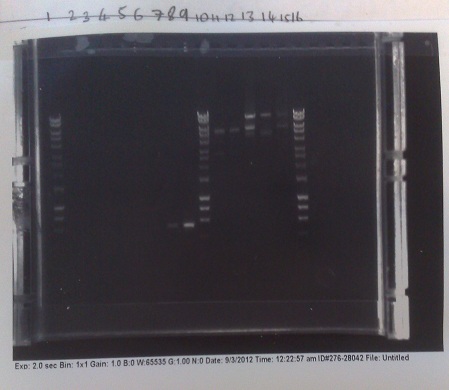
Verification of Mg2+ riboswitch construct by colony PCR
- Gibson primers used to amplify out riboswitch section of plasmid produced and transformed into e.coli to verify if riboswitch had inserted into PJS130.
- Band at ~550bp in lanes 8 and 9 (replicates) indicate that the riboswich has only been successfuly transformed into the second of the two colonies picked on the +8 plate, and not at all for the -8 construct.
- E.coli from strains made yesterday miniprepped to get plasmid DNA at high concentration. After colony PCR results, all DNA other than that of the successful colony discarded.
Verification of Mg2+ riboswitch construct by restriction digest
- BamH1 and Sal1 used to digest plasmid DNA produced from miniprep.
- Resultant fragment run on gel.
- Bands produced inconclusive, possibly because of too short an incubation time with restriction enzymes. Expected bands at ~4900bp and ~4500bp. Will re-run experiment in future to check results of colony PCR.
Week 11
Wednesday (05/09/12)

PCR of biobrick fragments and MGRS construct fragments.
- Fluoride and magnesium riboswitch genomic DNA amplified with PCR, along with vector fragments for turning the MGRS into a biobrick and for producing the magnesium riboswitch construct.
- PCR settings: Annealing temperature - 58 °C, Elongation step - 60 seconds.
- Genomic fragments worked well, but longer backbone fragments mostly failed.
- Extractions of DNA from these gels failed almost completely. Will try purifying directly from PCR products next time - results may not produce such a high purity of the precise DNA we want, but the increase in yield should outweigh this problem.
Gibson assembly of -8 MGRS construct
- Fragments gel extracted earlier assembled with Gibson assembly.
- Fragments used:
- 9kbp fragment from tube 13 used as backbone.
- Genomic fragments from tubes 6 + 7.
Transformation of E.coli with Gibson products
- Gibson products from earlier today used to transform e.coli.
- Resultant transformants plated out on 100 μg/ml ampicillin plates.
Week 12
Monday (10/09/12)
- Attempted to run PCR of the backbone for the biobricks once more. Settings: Annealing temperature: 56°C, elongation time: 30secs.
- After running on gel, saw that while the -8 vector amplification appears to be producing some correctly sized (~2kb) bands, the other two only seem to be forming many primer dimers. Analysis of the sequence of the prefix and suffix revealed a CG palindrome that appears to be causing self annealing. Inserts are not affected, as they have the palindrome at the 5' end of the primer.
- Will retry at higher temperatures tomorrow.
- Attempted Gibson assembly for the -8 magnesium riboswitch biobrick with the backbone fragments produced earlier today and the insert fragments produced yesterday.
Transformation of e.coli with Gibson products
- E.coli cells transformed with Gibson products produced earlier today.
- Transformants plated out on 25μ/ml chloramphenicol plates.
Restriction digest of magnesium riboswitch construct plasmids
- Colonies grown up from colonies produced from Gibson products made on Check!!! were miniprepped and plasmid DNA extracted.
- Restriction digest performed with Sal1 and BamH1. Expected fragment sizes: 4900bp and 4500bp. If plasmid lacks insert: 4900bp and 3950bp. These two possibilities should be distinguishable.
- Correct banding pattern seen for all colonies grown up, with both -8 and +8 construct. Shall verify identity further with colony PCR.
Thursday (13/09/12)
Magnesium Riboswitch Plate-Reader Assay
- Plate reader assay set up with magnesium riboswitch construct made over the last few days.
- Mg2+ concentrations used:
- 5μM, 10μM, 20μM, 50μM, 100μM, 200μM, 500μM, 1mM, 2mM, 5mM, 10mM, 20mM
- IPTG concentrations used:
- 0.05mM, 0.1mM, 0.2mM, 0.5mM, 1mM, 2mM, 5mM, 10mM
- Magnesium free medium B used as growth medium
- Results: fluorescence curves are constant, except for a distinct dip during the exponential phase of bacterial growth. After looking at various cultures under the fluorescence microscope and discussing the results with our advisors, we have decided that this is because the amino acids in the medium are fluorescing. Our bacteria are then degrading these, causing the dip observed. We will switch to M9 minimal medium for our assays in the future, as this does not autofluoresce.
Week 13
Week 14
Wednesday (26/09/12)
Miller Assay of Fluoride construct
- colonies picked from overnight and Miller assay run again as directed on protocols page
- 2 colonies each of E. coli, 168 strain, and crcB knockout strain Bacillus assayed
- Greater resolution at lower fluoride concentrations to test theory from previous assay
- Lack of filters meant that only A450 data recorderd
 "
"