Team:Penn/Targeting
From 2012.igem.org
| (2 intermediate revisions not shown) | |||
| Line 42: | Line 42: | ||
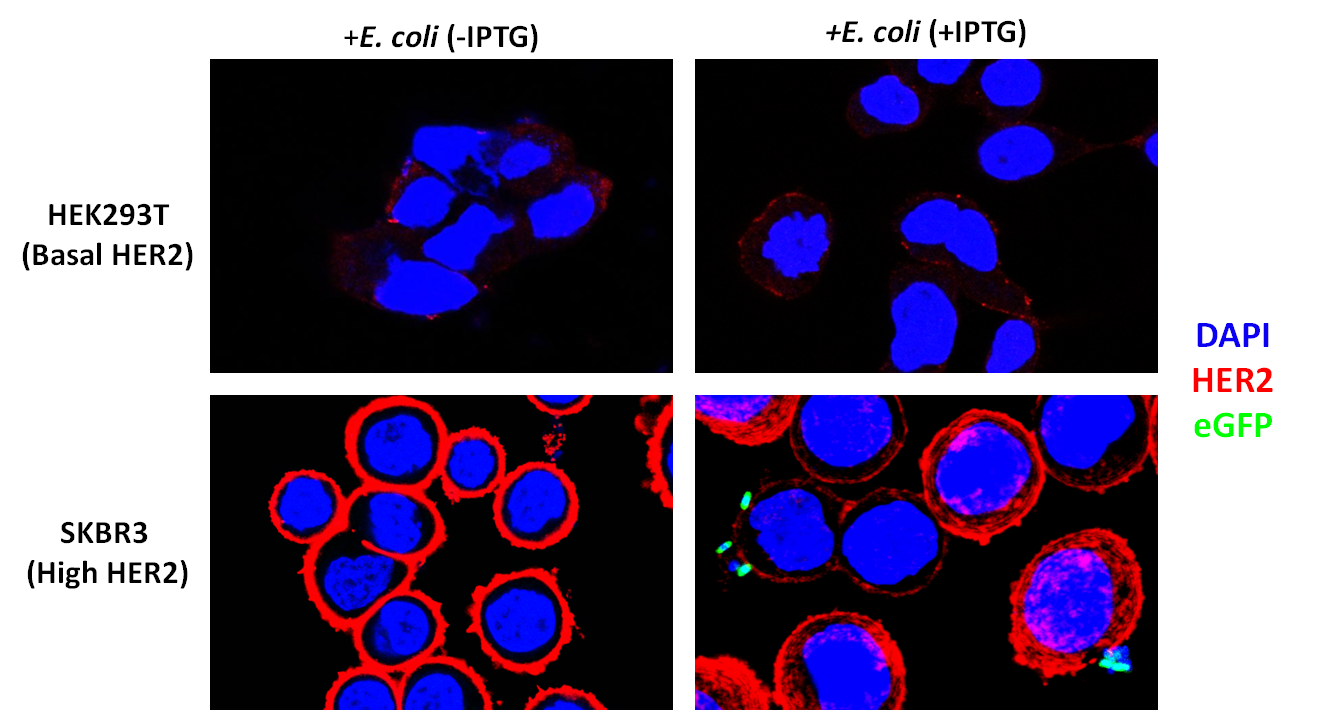
<div class="fig11"><b>Figure 3</b>: SKBR3 cells overexpress HER2 and DARPin-displaying bacteria bind to their surface. Red indicates HER2 (Alexa-Fluor 647). Blue indicates DAPI. Green indicates eGFP. Bacteria were co-incubated at a Multiplicity of Infection of 200:1 and washed three times after co-incubation followed by a 50ug/mL gentamicin protection assay.</p></div> | <div class="fig11"><b>Figure 3</b>: SKBR3 cells overexpress HER2 and DARPin-displaying bacteria bind to their surface. Red indicates HER2 (Alexa-Fluor 647). Blue indicates DAPI. Green indicates eGFP. Bacteria were co-incubated at a Multiplicity of Infection of 200:1 and washed three times after co-incubation followed by a 50ug/mL gentamicin protection assay.</p></div> | ||
| + | <br> | ||
| + | <br> | ||
| + | Taking a closer look at the SKBR3 + Induced Bacteria group, one can observe our eGFP labeled bacteria bound directly to the HER2 on SKBR3 cells (Figure 4) | ||
| + | <br> | ||
| + | <br> | ||
| + | <div align="center"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/9/94/SKBR3_closeup_binding.png" width="900" height="494.6" /></div> | ||
| + | <div class="fig11"><b>Figure 4</b>: Direct binding of INPNC-DARPin expressing, eGFP-labeled bacteria to SKBR3 cells.</p></div> | ||
| - | In summary, we demonstrated surface display of DARPin H10-2-G3 for the first time and have | + | |
| + | |||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <b>Conclusion</b> | ||
| + | <p style="color:black;text-indent:30px;"> | ||
| + | In summary, we demonstrated surface display of DARPin H10-2-G3 for the first time and have demonstrated that our DARPin-displaying bacteria can target cancer cells <i>in vitro</i>. We also developed a generalized surface display BioBrick for any iGEM team to use.<br /> </div></div> | ||
</html> | </html> | ||
Latest revision as of 02:59, 27 October 2012
After verifying that the DARPin was displayed on the cell surface, we sought to test whether it could allow binding to HER2-overexpressing cancer cells. We were generously provided SKBR3 cells by the lab of Matthew Lazzara. These cells are derived from breast cancer cells and over-express HER2. We also obtained Human Embryonic Kidney (HEK) 293T cells, which express a low amount of HER2, to act as controls.
To verify that our SKBR3 cells overexpressed HER2, we immunostained for HER2 (primary anti-Neu Mouse Igg, Santa Cruz Biotechnology and secondary donkey anti-mouse Alexa-Fluor 647 conjugate, Jackson Immunoresearch) and visualized the cells on a confocal microscope. As expected, HER2 was overexpressed in SKBR3 cells (Figure 1).

Figure 1

Figure 2
To assay whether our DARPin-displaying E. coli bound to SKBR3 cells preferentially, we conducted experiments in which our bacteria were co-incubated with SKBR3 or HEK293T cells. The DARPin-displaying bacteria were co-transformed with constitutively expressed eGFP (pACBB-eGFP plasmid) for easy visualization. The goal was to show SKBR3-specific binding of eGFP labeled bacteria, and only when they were induced with IPTG to produce INPNC-DARPin. Preliminary results are exciting and show preferential binding to SKBR3 cells. In culture plates with low-HER2 HEK293T cells, no green bacteria were observed bound to the cells. IPTG-induced bacteria bound selectively to SKBR3 cells and we visualized this binding by confocal microscopy (Figure 3).
Taking a closer look at the SKBR3 + Induced Bacteria group, one can observe our eGFP labeled bacteria bound directly to the HER2 on SKBR3 cells (Figure 4)

Conclusion
In summary, we demonstrated surface display of DARPin H10-2-G3 for the first time and have demonstrated that our DARPin-displaying bacteria can target cancer cells in vitro. We also developed a generalized surface display BioBrick for any iGEM team to use.
 "
"

