Team:SJTU-BioX-Shanghai
From 2012.igem.org
m |
Huanan1991 (Talk | contribs) |
||
| (113 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{Template:12SJTU_main_header}} | |
| - | + | ||
<html> | <html> | ||
| - | < | + | <body> |
| - | < | + | <!-- =============================================================================== --> |
| - | + | ||
| - | < | + | <table class="mainouter" width="1012" cellspacing="0" cellpadding="5" align="center"> |
| - | <div id=" | + | <tbody> |
| - | + | <tr><td class="pumpkin"> | |
| - | </ | + | </td></tr> |
| - | < | + | <tr><td class="text" align="center"> |
| - | + | <table class="main" width="1010" border="0" cellspacing="0" cellpadding="0"><tbody> | |
| - | </ | + | <tr><td class="embedded"> |
| - | </div> | + | <div> |
| - | </ | + | <ul class="semiopaquemenu navipages" id="lavalamp"> |
| + | <li class="current"><a href="/Team:SJTU-BioX-Shanghai" class="selected">Home</a></li> | ||
| + | <li><a href="/Team:SJTU-BioX-Shanghai/Project">Project</a></li> | ||
| + | <li><a href="/Team:SJTU-BioX-Shanghai/Parts">Parts</a></li> | ||
| + | <li><a href="/Team:SJTU-BioX-Shanghai/Team">Team</a></li> | ||
| + | <li><a href="/Team:SJTU-BioX-Shanghai/Notebook/protocol">Notebook</a></li> | ||
| + | <li><a href="/Team:SJTU-BioX-Shanghai/Consideration/human">Human Practice</a></li> | ||
| + | <li><a href="/Team:SJTU-BioX-Shanghai/Consideration/safety">Consideration</a></li> | ||
| + | </ul> | ||
| + | </div> | ||
| + | |||
| + | </td></tr> | ||
| + | </tbody></table> | ||
| + | </td></tr> | ||
| - | <!-- | + | <tr><td align="center" class="outer"> |
| + | <table width="1010" class="main" border="0" cellspacing="0" cellpadding="0"><tbody> | ||
| + | <tr><!-- abstract --> | ||
| + | <td> | ||
| + | <div id="abstract"> | ||
| + | <div id="abstract-title"></div> | ||
| + | <div id="abstract-content"> | ||
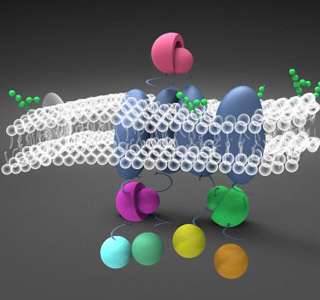
| + | <p>This year, SJTU-BioX-Shanghai iGEM team has build a "factory" on E.coli's membrane, where enzyme assemblies can be manipulated so that biochemical reactions can be accelerated and further controlled. </p> | ||
| + | <p>We aimed at constructing a set of protein assemblies on E.coli inner membrane as scaffold carrying various enzymes. Distinct from previous synthetic scaffold system, our device limits the reaction space to a two-dimensional surface. In such system, the membrane functions as an extensive scaffold for proteins to anchor without limitation of scaffold amount. Membrane as scaffold also has privilege in receiving external and internal regulating signals. Based on Membrane Scaffold, we built two universal devices: Membrane Accelerator and Membrane Rudder.</p> | ||
| + | <p>In Membrane Accelerator device, by gathering enzymes on membrane, production of fatty acid was enhanced by more than 24 fold, which has a promising application prospect in biofuel production. We also proposed a new direction for application of scaffold-based accelerator: Biodegradation. </p> | ||
| + | <p>In Membrane Rudder device, we changed the direction of Violacein synthetic pathway through external signal. </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr><!-- design of membrane system--> | ||
| + | <td style="border-top-width:40px;"> | ||
| + | <div id="design"> | ||
| + | <div id="design-title"></div> | ||
| + | </div> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr><!-- 3 entrys --> | ||
| + | <td style="border-top-width:20px;" align="center"> | ||
| + | <div class="main-entry" style="background-color:#ffb000; margin-right:26px;"> | ||
| + | <div class="entry-title"> | ||
| + | <img src="/wiki/images/2/2e/12SJTU_title_1.png"> | ||
| + | </div> | ||
| + | <div class="entry-content"> | ||
| + | <img src="/wiki/images/3/34/12SJTU_entry1.jpg" style="width:320px"></img> | ||
| + | </div> | ||
| + | <a href="/Team:SJTU-BioX-Shanghai/Project/project2.2"><div id="entry-cover1"> | ||
| + | <p>Membrane Accelerator could help substrates flow by decreasing distance that intermediates have to travel between enzymes. Besides, restricted reaction space on membrane could increase the local products concen- tration near membrane, thus to facilitate the exportation of final products </p> | ||
| + | <p>We successfully increased the yield of fatty acids by 24 fold through building a fatty-acid-producing Mem- brane Accelerator.</p> | ||
| + | </div></a> | ||
| + | </div> | ||
| + | <div class="main-entry" style="background-color:#4fb1ca; margin-right:26px;"> | ||
| + | <div class="entry-title"> | ||
| + | <img src="/wiki/images/6/6b/12SJTU_title_2.png"> | ||
| + | </div> | ||
| + | <div class="entry-content"> | ||
| + | <img src="/wiki/images/a/a4/12SJTU_entry2.jpg"></img> | ||
| + | </div> | ||
| + | <a href="/Team:SJTU-BioX-Shanghai/Project/project2.3"><div id="entry-cover2"> | ||
| + | |||
| + | <p>Previous synthetic scaffold con- trolling metabolic flux all focused on biosynthesis. Now we propose a new direction in the application of syn- thetic scaffold: accelerating biodegradation pathways. </p> | ||
| + | <p>Natural biodegradation is a very slow process but indispensible in environment restoration. Membrane Accelerator is expected to increase rate of biodegrada- tion pathway sharply due to its multiple privileges.</p> | ||
| + | </div></a> | ||
| + | </div> | ||
| + | <div class="main-entry" style="background-color:#fe88b8;"> | ||
| + | <div class="entry-title"> | ||
| + | <img src="/wiki/images/d/d7/12SJTU_title_3.png"> | ||
| + | </div> | ||
| + | <div class="entry-content"> | ||
| + | <img src="/wiki/images/2/24/12SJTU_entry3.jpg"></img> | ||
| + | </div> | ||
| + | <a href="/Team:SJTU-BioX-Shanghai/Project/project2.1"><div id="entry-cover3"> | ||
| + | |||
| + | <p>Dynamically and artificially regulating the direction of biochemical pathway has remained a challenge to all. Based on Membrane Scaffold, we achieved this goal through controlling aggregation state of crucial enzymes in branched reactions with light. This novel device is called Membrane Rudder.</p> | ||
| + | <p>We then further connected the post-translational control system (Membrane Rudder) to genetic circuits and expanded its potential signal pool.</p> | ||
| + | </div></a> | ||
| + | </div> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
| + | <tr><td style="border-top-width:40px;"><!-- achievements--> | ||
| + | <div id="achievements"> | ||
| + | <div id="achievements-title"></div> | ||
| + | <div id="achievements-content"> | ||
| + | |||
| + | <p><strong><img src="/wiki/images/7/73/12SJTU_r.png"><font face="Cambria, serif " size="4"> The honors we have won:</font></strong> Regional Winner, Best New BioBrick Part or Device Engineered and Gold Medal at Asia Jamboree; Advanced to World Championship.</p> | ||
| + | <p><strong><img src="/wiki/images/7/73/12SJTU_r.png"><font face="Cambria, serif " size="4"> The breakthrough we made:</font></strong> Redefinition of scaffold in Synthetic Biology by recruiting <i>E.coli’</i>s inner membrane as a natural two-dimensional scaffold. </p> | ||
| + | <p><strong><img src="/wiki/images/7/73/12SJTU_r.png"><font face="Cambria, serif " size="4"> The system we built:</font></strong> 6 membrane proteins orderly organized on the inner membrane of <i>E.coli</i>, the feasibility of which has been confirmed by fluorescence complementation assay and biosynthesis experiment.</p> | ||
| + | <p><strong><img src="/wiki/images/7/73/12SJTU_r.png"><font face="Cambria, serif " size="4"> Device 1 we created – Membrane Accelerator:</font></strong> A universal tool that serves to accelerate biochemical reactions in <i>E.coli</i>; Rate of fatty acids synthesis was increased by 24 fold compared to wild-type <i>E.coli </i>and 9 fold compared to <i>E.coli</i> overexpressing cytoplasmic enzymes.</p> | ||
| + | <p><strong><img src="/wiki/images/7/73/12SJTU_r.png"><font face="Cambria, serif " size="4"> Device 2 we created – Membrane Rudder:</font></strong> A universal tool used to dynamically and artificially control biochemical reactions in <i>E.coli</i>; the direction of Violacein and Deoxyviolacein synthetic pathway was successfully switched through changing blue light signal. </p> | ||
| + | <p><strong><img src="/wiki/images/7/73/12SJTU_r.png"><font face="Cambria, serif " size="4"> New direction we proposed:</font></strong> The application of scaffold system in accelerating biodegradation pathway by using Membrane Accelerator.</p> | ||
| + | <p><strong><img src="/wiki/images/7/73/12SJTU_r.png"><font face="Cambria, serif " size="4"> Parts we submitted:</font></strong> 42 well-characterized parts that could either be used directly or serve as a universal tool readily for potential scientific or engineering use.</p> | ||
| + | <p><strong><img src="/wiki/images/7/73/12SJTU_r.png"><font face="Cambria, serif " size="4"> A club we established – BioCraft:</font></strong> The headquarter of our human practice programs, having come a long way in propagandizing Synthetic Biology and iGEM. Warmly-received activities have been held in and outside the campus. Several celebrities in different fields have shown support for us, laying a cornerstone for our future development.</p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </td></tr> | ||
| + | |||
| + | <tr><!-- team & video --> | ||
| + | <td style="border-top-width:40px;"> | ||
| + | <div id="team"> | ||
| + | <div id="team-title"></div> | ||
| + | <a href="/Team:SJTU-BioX-Shanghai/Team/members"><div id="team-content"> | ||
| + | <img src="/wiki/images/b/ba/12SJTU_home_Team.jpg"></img> | ||
| + | </div></a> | ||
| + | </div> | ||
| + | <div id="video"> | ||
| + | <iframe width="480" height="400" src="http://www.youtube.com/embed/PkE9MtApxAE" frameborder="0" allowfullscreen></iframe> | ||
| + | </div> | ||
| + | </td></tr> | ||
| + | |||
| + | <tr><td style="border-top-width:40px;"> | ||
| + | <div id="sponsors"> | ||
| + | <div id="sponsors-title"></div> | ||
| + | <div id="sponsors-content"> | ||
| + | <table class="sponsorstable" cellspacing="20" ;="" align="center"><tbody> | ||
| - | + | <tr> | |
| - | + | <td> | |
| - | == | + | <a href="http://www.sjtu.edu.cn/"> |
| + | <img src="/wiki/images/6/6f/12SJTU_SJTU.jpg" style="width: 140px;"> | ||
| + | </a></td> | ||
| + | <td> | ||
| + | <a href="http://www.lib.sjtu.edu.cn/"> | ||
| + | <img src="/wiki/images/e/ee/12SJTU_lib.jpg" style="width: 140px;"> | ||
| + | </a></td> | ||
| + | <td> | ||
| + | <a href="http://www.bio-x.cn"> | ||
| + | <img src="/wiki/images/1/1a/12SJTU_biox.png" style="width: 140px;"> | ||
| + | </a></td> | ||
| + | <td> | ||
| + | <a href="http://life.sjtu.edu.cn/"> | ||
| + | <img src="/wiki/images/5/5c/12SJTU_LS.png" style="width: 140px;"> | ||
| + | </a></td> | ||
| + | <td> | ||
| + | <a href="http://www.huaqiaofoundation.org/"> | ||
| + | <img src="/wiki/images/6/68/12SJTU_HP3.1.jpg" style="width: 140px;"> | ||
| + | </a></td> | ||
| + | </tr> | ||
| - | + | <tr> | |
| - | + | <td> | |
| - | + | <a href="http://www.lifetechnologies.com"> | |
| + | <img src="/wiki/images/d/de/12SJTU_Life_Technology.jpg" style="width: 140px;"> | ||
| + | </a></td> | ||
| + | <td> | ||
| + | <a href="http://www.genscript.com/"> | ||
| + | <img src="/wiki/images/5/55/12SJTU_genscript_logo.jpg" style="width: 140px;"> | ||
| + | </a></td> | ||
| + | <td> | ||
| + | <a href="http://www.dnbios.com/"> | ||
| + | <img src="/wiki/images/a/af/12SJTU_DNBIO.jpg" style="width: 140px;"> | ||
| + | </a></td> | ||
| + | <td> | ||
| + | <a href=""> | ||
| + | <img src="/wiki/images/5/55/12SJTU_shenglianshengwu.jpg" style="width: 140px;"> | ||
| + | </a></td> | ||
| + | <td> | ||
| + | <a href=""> | ||
| + | <img src="/wiki/images/f/f0/12SJTU_SHIYI.jpg" style="width: 140px;"> | ||
| + | </a></td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | </div> | ||
| + | </div> | ||
| + | </td></tr> | ||
| + | <!-- !!!!!!!!!!!!!! --> | ||
| + | </tbody></table> | ||
| + | </td></tr> | ||
| + | </tbody></table> | ||
| + | <br> | ||
| + | |||
| - | + | <div id="toolBackTo" class="back-to" > | |
| + | <a class="backtotop" href="#top" onclick="window.scrollTo(0,0);return false;">To Top | ||
| + | |||
| + | </a> | ||
| + | </div> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | <div align="center" id="footer"> (c) <a href="" target="_self">SJTU_BioX_iGEM2012</a> Powered by huanan1991</div> |
| - | + | </div> | |
| - | + | </body> | |
| - | + | </html> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 03:58, 27 October 2012
| ||||||
|
 "
"





 The honors we have won: Regional Winner, Best New BioBrick Part or Device Engineered and Gold Medal at Asia Jamboree; Advanced to World Championship.
The honors we have won: Regional Winner, Best New BioBrick Part or Device Engineered and Gold Medal at Asia Jamboree; Advanced to World Championship.









