Team:SJTU-BioX-Shanghai/Project/project2.1
From 2012.igem.org
Zhiantinglan (Talk | contribs) (→Membrane Rudder - Violacein & Deoxyviolacein synthetic pathway) |
Zhiantinglan (Talk | contribs) (→Membrane Rudder Sensing Light Signal) |
||
| (53 intermediate revisions not shown) | |||
| Line 24: | Line 24: | ||
<td valign="top" width="750"> | <td valign="top" width="750"> | ||
__NOTOC__ | __NOTOC__ | ||
| - | =Membrane Rudder - Violacein & Deoxyviolacein | + | =Membrane Rudder <br><br> - Violacein & Deoxyviolacein Synthetic Pathway= |
{{Template:12SJTU_part_summary_head}} | {{Template:12SJTU_part_summary_head}} | ||
| - | *'''State of the art''' | + | |
| + | *'''State of the art''' | ||
Dynamically and artificially regulating the direction of biochemical pathway ''in vivo'' has remained a challenge for scientists. We are now to achieve this goal through controlling the aggregation state of different enzymes in branched biological reactions based on Membrane Scaffold. We named this universal device ''Membrane Rudder''. | Dynamically and artificially regulating the direction of biochemical pathway ''in vivo'' has remained a challenge for scientists. We are now to achieve this goal through controlling the aggregation state of different enzymes in branched biological reactions based on Membrane Scaffold. We named this universal device ''Membrane Rudder''. | ||
| Line 42: | Line 43: | ||
*'''Achievements''' | *'''Achievements''' | ||
| - | + | ''Direction switched'' successfully in Violacein synthetic pathway by light signal | |
| - | + | ''8-fold yields decrease'' of side product deoxychromaviridans with ''Membrane Rudder'' compared to group with free cytoplasmic enzyme | |
| - | + | ''A novel device constructed'' that connects post-translational Membrane Scaffold system to genetic circuits by recruiting RNA as a controlling signal. | |
{{Template:12SJTU_part_summary_foot}} | {{Template:12SJTU_part_summary_foot}} | ||
| Line 52: | Line 53: | ||
==Background== | ==Background== | ||
| - | + | Dynamically and artificially regulating the direction of biochemical pathway ''in vivo'' has remained a challenge for scientists. Based on Membrane Scaffold, we are now trying to achieve this goal through controlling the aggregation state of different enzymes in branched biological reactions. We named this universal device ''Membrane Rudder''. Theoretically this device could sense a large amount of signals. We chose blue light and rationally designed RNA molecule D0 as controlling signal. Branched violacein & deoxyviolacein synthetic pathway is recruited to verify the feasibility of ''Membrane Rudder''. | |
| - | There are three branches in | + | There are three branches in violacein & deoxyviolacein synthetic pathway. We could identify amount of final products through high-performance liquid chromatography (HPLC). The change of products proportion will demonstrate the efficiency of direction alteration. |
| - | |||
| - | |||
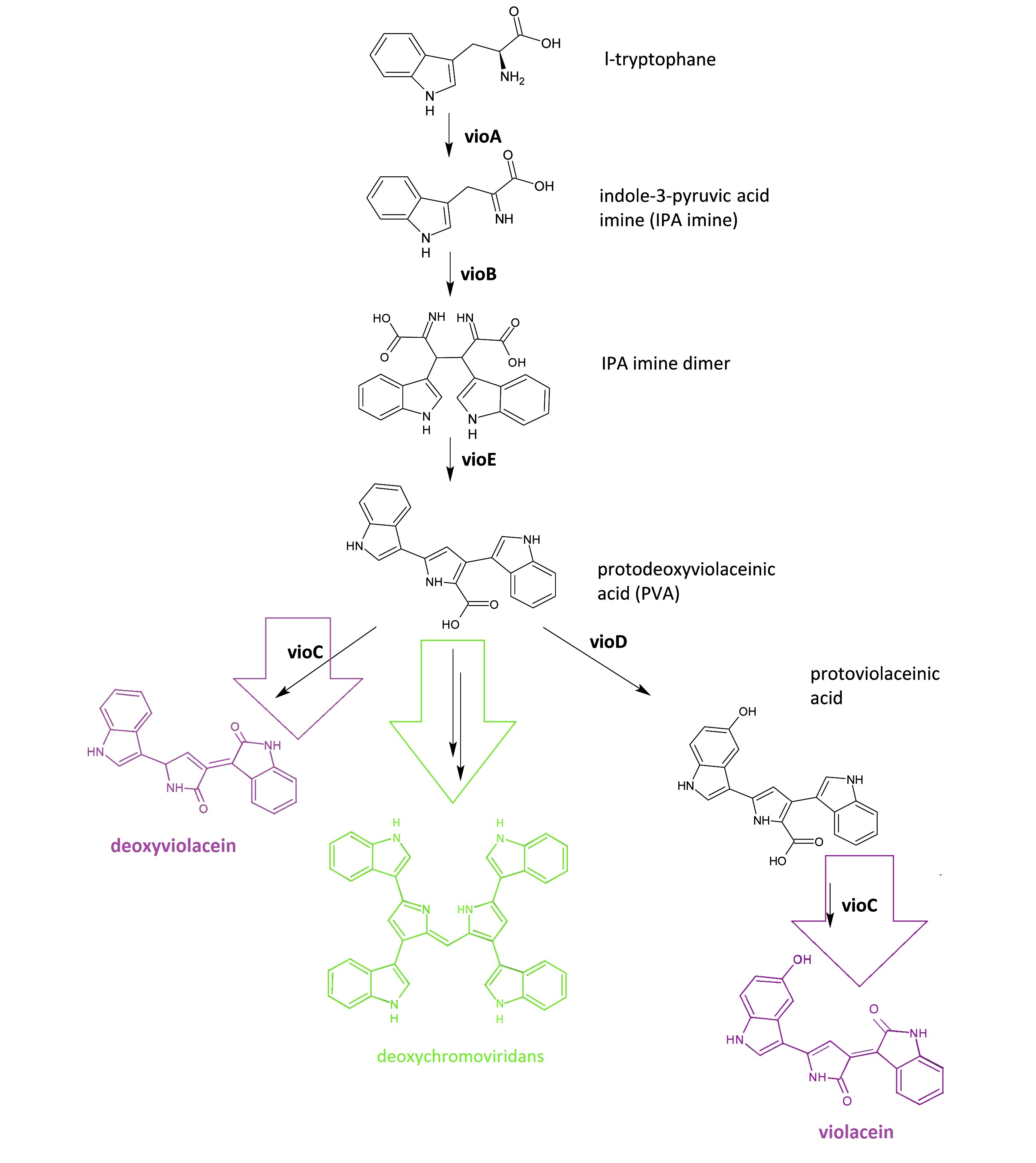
| - | + | ===Violacein & Deoxyviolacein Biosynthetic Pathway=== | |
| - | + | ::[[Image:Figure1-SJTU violacein-pathway.png|thumb|600px|center|''Fig.1:'' :Details of violacein & deoxyviolacein biosynthetic pathway. The left pathway painted in purple indicates the synthetic pathway of purple pigment deoxyviolacein.The right pathway painted in violet indicates the synthetic pathway of purple pigment violacein. The green pathway indicates the nonenzymatic production of green pigment deoxychromoviridans.]] | |
| - | The | + | The violacein & deoxyviolacein biosynthetic pathway includes 5 key enzymes which work in sequence: Vio A, B, C, D and E. VioA, a flavoenzyme and VioB, a heme protein, work together to oxidize and dimerize tryptophen into IPA imine dimer. Then VioE induces an indole rearrangement, producing prodeoxyviolacein acid, also known as PVA. In a word, enzyme VioA, VioB and VioE convert tryptophen into PVA. Then this biosynthetic pathway is about to branch. |
| - | + | In ''E.coli'', an additional side reaction could occur, and PVA is converted into a green pigment called deoxychromoviridans. | |
| - | + | ||
| - | + | The last two enzymes, VioC and VioD are flavin-dependent oxygenases. VioC alone transforms PVA into a purple pigment called deoxyviolacein. VioC could also act cooperatively with VioD. VioD hydroxylates 5-position indole ring, and then the other 2-position indole ring is processed by VioC to create the oxindole. In this way, violacein is produced. | |
| - | + | ||
| - | + | ==Design of Experiment== | |
| - | + | We tried to create ''Membrane Rudder'' controlled by light signal and RNA signal to switch direction of biochemical reaction dynamically and artificially in ''E.coli''. | |
| - | + | ===Membrane Rudder Sensing Light Signal=== | |
| + | Light is an intriguing signal to regulate E.coli activity because it is easy to obtain, highly tunable and nontoxic. A metabolite-coupled light-switchable system could be quite fascinating. | ||
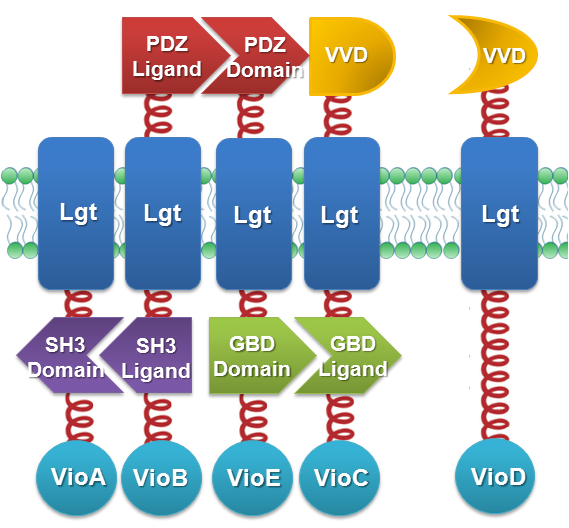
| - | + | We constructed our device as demonstrated in ''Fig.2''. In light-sensing ''Membrane Rudder'', VioA, VioB, VioE and VioC with interacting Membrane Anchors constitutively aggregate. [https://2012.igem.org/Team:SJTU-BioX-Shanghai/Project/project1.2 VVD], a photoreceptor of ''Neurospora crassa'', can form dimer in the presence of blue light and disassociate as light is off. When blue light is present, VioD with VVD-coupled Membrane Anchor would aggregate with assembly of VioA, B, C and E, making violacein the dominant final product. But when blue light is off, VioD with VVD coupled Membrane Anchor will disassociate with VioA, B, C and E. Thus the biosynthetic pathway for deoxyviolacein is switched on. | |
| - | + | [[File:12SJTU_VVD_Construction.png|center|500px|thumb|''Fig.2'' :Construction details of light-sensing ''Membrane Rudder''. VVD acts as a blue light sensor.]] | |
| - | + | For VioB and VioE only function normally in dimeric state, free VioB and free VioE were coexpressed with membrane anchored VioA, B, C, D and E (''Fig.2'')to ensure the normal function of the whole system. | |
| - | + | Bacteria in experimental group were induced at a L-Arabinose concentration of 0.1%. One group of bacteria expressing full set of light-sensing ''Membrane Rudder'' is incubated under blue light. The other group of bacteria expressing full set of light-sensing ''Membrane Rudder'' are incubated in the dark. In each comparative group, bacteria prepared for light induction and dark incubation are taken from the same sample. | |
| - | + | The BioBrick Part VVD-MA5-vioC and VVD-MA6-vioD is [http://partsregistry.org/Part:BBa_K771205 Part:BBa_K771205] and [http://partsregistry.org/Part:BBa_K771206 Part:BBa_K771206], respectively. For MA2-VioA, MA3-VioB, MA4-VioE, the corresponding part is [http://partsregistry.org/Part:BBa_K771201 Part:BBa_K771201], [http://partsregistry.org/Part:BBa_K771202 Part:BBa_K771202], [http://partsregistry.org/Part:BBa_K771203 Part:BBa_K771203], respectively. | |
| + | |||
| + | ===Membrane Rudder Sensing RNA Signal=== | ||
| + | |||
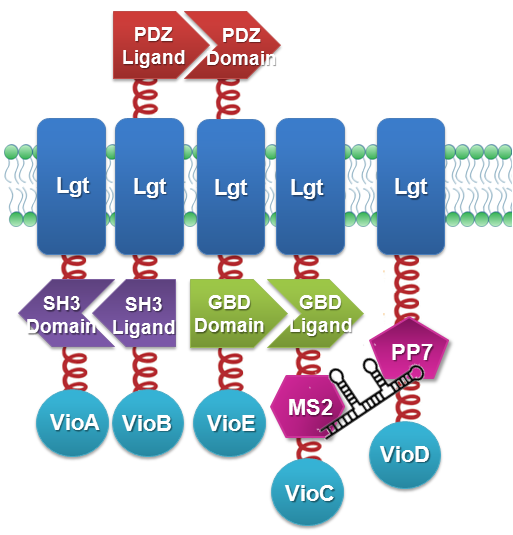
| + | Membrane Accelerator and Membrane Rudder are both post-translational controlling device to regulate metabolic flux of the host cell. To connect this relatively isolated post-translational control system to genetic circuits, we employed RNA signal, which is present in cytoplasm. Rationally designed RNA D0 with MS2 and PP7 aptamer domain is recruited. When RNA molecule with this two aptamer domains is present in cells, their cognate aptamer binding proteins can thus aggregate together. Furthermore, if we place RNA D0 (with PP7 and MS2 aptamer domains) under various promoters regulated by different signals, approaches to induce dimerization would be expanded sharply. Thus, Membrane Rudder could sense much more signals. | ||
| + | |||
| + | We constructed our device as demonstrated in ''Fig.3''. In RNA-sensing ''Membrane Rudder'', VioA, VioB, VioE and VioC with interacting Membrane Anchors constitutively aggregate. [https://2012.igem.org/Team:SJTU-BioX-Shanghai/Project/project1.2 RNA D0], can aggregate with RNA aptamer binding protein MS2 and PP7. So when RNA D0 is present, VioD with MS2-coupled Membrane Anchor would aggregate with assembly of VioA, B, C and E, making violacein the dominant final product. But when RNA D0 is absent, VioD with MS2-coupled Membrane Anchor will disassociate with VioA, B, C and E. Thus the biosynthetic pathway for deoxyviolacein is switched on. | ||
| + | |||
| + | [[File:12SJTU_RNA_construction.png|center|500px|thumb|''Fig.3'' :Construction details of RNA-sensing ''Membrane Rudder''. RNA aptamer binding protein MS2 and PP7 act as an RNA signal sensors.]] | ||
| + | |||
| + | Free VioB and free VioE were coexpressed with membrane anchored VioA, B, C, D and E to ensure the normal function of the whole system. | ||
| + | |||
| + | Bacteria in experimental group were induced at a L-Arabinose concentration of 0.1%. One group of bacteria expressing full set of RNA-sensing ''Membrane Rudder'' are incubated under blue light. The other group of bacteria expressing full set of RNA-sensing ''Membrane Rudder'' are incubated in the dark. In each comparative group, bacteria prepared for light induction and dark incubation are taken from the same sample. | ||
==Results and Discussion== | ==Results and Discussion== | ||
| - | === | + | ===Membrane Rudder Sensing Light Signal=== |
| - | + | ||
| - | + | '''Overview''' | |
| - | + | ||
| - | + | By attaching Enzymes VioA, B, C, D and E to Membrane Anchors as shown in ''Fig.2'', we managed to change the proportion of final products by switching on or off blue light signal. When blue light is on, VioA, B, C, D and E aggregate together, making violacein the dominant final product. When blue light is off, VioD with Membrane Anchor will disassociate with VioA, B, C and E. Thus the biosynthetic pathway for deoxyviolacein is switched on. | |
| - | + | Moreover, a significant decrease of side products such as deoxychomoviridans was observed when ''Membrane Rudder'' system was present. | |
| - | + | ||
| - | |||
| - | |||
| - | + | '''Direction Alteration''' | |
| - | + | Altering direction of violacein & deoxyviolacein synthetic pathway can be realized by dynamically controlling the aggregation state of crucial enzymes. Through the blue light induction, all five enzymes aggregate together. Thus branch pathway producing deoxyviolacein by VioA, B, E and C is inhibited because PVA is more preferably hydroxylated by VioD. On the other hand, as long as blue light is absent from the bacteria, VioD will disassociate with assembling VioA, B, E and C, leading to the production of deoxyviolacein. | |
| - | + | [[Image:P1000998.JPG|thumb|400px|center|''Fig.4'' :Collections of different peaks in HPLC results, ready to undergo MS for molecule construction verification.]] | |
| + | |||
| + | We ran an HPLC (SHIMADZU LC-20AP, C18 reversed column) test with the purple samples extracted from the bacteria culture. Then we ran mass spectrometry (Thermo Ultra GC-ISQ) test and thus further confirmed the molecular constitution of main peaks in HPLC results. | ||
| + | |||
| + | The HPLC results perfectly fit our prediction. | ||
| + | |||
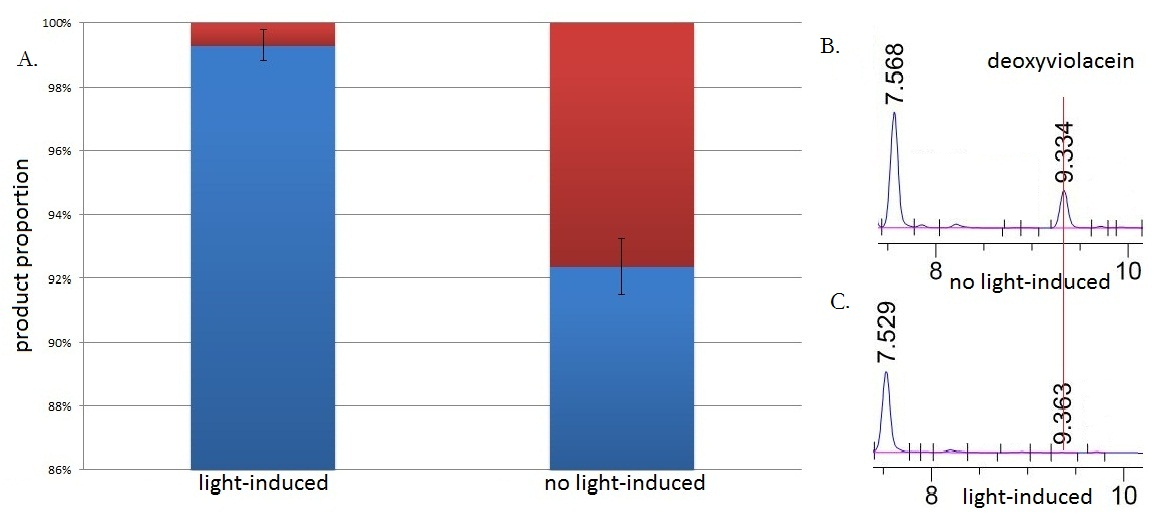
| + | [[Image:12SJTU_vio_propotion4.jpg|thumb|700px|center|''Fig.5'' :HPLC result (right) and the ratio(left) of deoxyviolacein in sample group with blue light induction and group without blue light induction. Red column stands for yield of deoxyviolacein; Blue column stands for yield of violacein. Fig. B shows the HPLC result from sample without blue light induction. Fig. C shows the HPLC result from sample with from sample under blue light induction.The HPLC results show that the peak of deoxyviolacein appears at about 9 minutes after the injection. The peak of deoxyviolacein is identified according to previously published research. Results indicate that the amount of the deoxyviolacein from the sample without blue light induction is tremendously larger than that from light induced sample]] | ||
| + | |||
| + | |||
| + | Under the induction of blue light, production of deoxyviolacein was inhibited (to almost 0%) while most products were violacein, which is predicted above. Without blue light induction, however, the pathway leading to the production of deoxyviolacein was turned on and thus considerable amount of deoxyviolacein was produced. All results confirmed the feasibility of our light-sensing ''Membrane Rudder''. Besides, it indicated that by replacing signal sensors and functioning enzymes, we could regulate direction of various branched reactions through diversified signals. | ||
| + | <br> | ||
| - | + | '''Decrease in Side Products''' | |
| - | + | ||
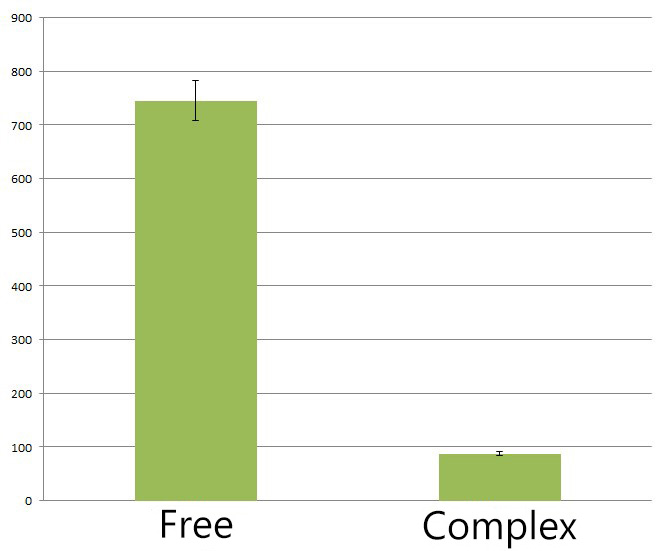
| - | + | We also found membrane anchored enzymes could help to reduce the amount of side-product(deoxychromaviridans) by 8 fold compared to diffusing cytoplasmic enzymes. This is because rationally designed enzyme assembly facilitate and improve the metabolic flux of biosynthesis. Intermediate PVA is more preferably processed by adjacent membrane anchored VioD or VioC rather than converted into deoxychromaviridans. Actually this is one of the advantages of Membrane Scaffold. | |
| - | + | [[Image:12SJTU_Virdrans_before_after.jpg|thumb|400px|center|''Fig.5'' :The HPLC peak area of deoxychromoviridans from sample with free VioABCDE(denoted as ''Free'') and membrane anchored VioABCDE(denoted as Complex). Construction detail of membrane anchored VioABCDE is shown in ''Fig.2''. Bacteria are incubated under light for 6 hours in ''Complex'' group.]] | |
==Reference== | ==Reference== | ||
Latest revision as of 03:55, 27 October 2012
| ||
|
 "
"