Team:Potsdam Bioware/Lab/Labjournal/October
From 2012.igem.org
(→12.10.2012) |
(→2012-10-20) |
||
| (40 intermediate revisions not shown) | |||
| Line 42: | Line 42: | ||
<Br> | <Br> | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-11</p>=== | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;"> Purification of Retrafo plasmids</p> | ||
| + | |||
| + | |||
| + | <b>Investigators:</b> Rico | ||
| + | |||
| + | |||
| + | <b>Results:</b> inoculation was not successful | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-26</p>=== | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;"> Purification of transfected wt AID, modified AID, TAL-AID plasmids cotransfected with small antibody construct and transformation</p> | ||
| + | |||
| + | |||
| + | <b>Investigators:</b> Rico, Stefan, Tom | ||
| + | |||
| + | |||
| + | <b>Method:</b> transfected cells were lysed, plasmids purified and transformed into ''E. coli cells'' | ||
==Antibody== | ==Antibody== | ||
| Line 54: | Line 74: | ||
<b>Topic: </b>cell-culture <br> | <b>Topic: </b>cell-culture <br> | ||
<br> | <br> | ||
| + | * passaging of CHO w Zeocin | ||
* passaging of HT1080 | * passaging of HT1080 | ||
| - | * | + | * passaging of HEK AAV 293 |
| - | + | * passaging of HeLa | |
| - | * | + | |
| - | * | + | |
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;"> Topic: planning new primer for integration of new RAGE-transmembrane domain and RAGE-signalpeptide </p> | ||
| + | |||
| + | <b>Investigator:</b> Sascha <br> | ||
| + | |||
| + | <b>Materials and Methods:</b> Geneious <br> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;"> Topic: Primer design and ordering for integration of RAGE-transmembrane domain into scFVconstruct and nanobody-geneart construct</p> | ||
| + | |||
| + | <b>Investigator:</b> Sascha <br> | ||
| + | |||
| + | <b>Materials and Methods:</b> Geneious <br> | ||
| + | |||
| + | <b>Results:</b><br> | ||
| + | |||
| + | * scFv:startprimer-fw | ||
| + | ** GCCTCTAGAGCTAGCATGGCAGC | ||
| + | * rvp:scfv+overhang to TEV/TMD | ||
| + | **CGCCCAAGATTCCCAGGGCCAGGGCGAGGGTGCCGCTTCCGCCGCCACCGCTTCCCCCTTGAAAATATAAATTCTCTCCAGATCCCCGTTT | ||
| + | GATCTCCAGTTCTGTCC | ||
| + | * fwp:nterm. of yfp to TMD | ||
| + | ** CCCTGGGAATCTTGGGCGGCCTGGGGACCGCTGCCCTCTTGATCGGCGTGATCCTGTGGCAGAGAAGGTCTGGAGTGAGCAAGGGCGAG | ||
| + | GAGC | ||
| + | * scFv:endprimer-rv | ||
| + | ** GAGCTGCAGCGGCCG | ||
| + | * rage:startprimer-fw_nano/Fc | ||
| + | ** CTTGAATTCGCGGCCG | ||
| + | * rage-rv:Fc to rageTMD | ||
| + | ** CGCCCAAGATTCCCAGGGCCAGGGCGAGGGTGCCGCTTCCTAATAACTTCGTATAATGTATGCTATACGAAGTTATCCC | ||
| + | * rage-fwp:mcherry/rageTMD | ||
| + | ** CCCTGGGAATCTTGGGCGGCCTGGGGACCGCTGCCCTCTTGATCGGCGTGATCCTGTGGCAGAGAAGGGGCTCCATGGTGTCCAAGG | ||
| + | * rage:endprimer in mcherry/loxp | ||
| + | ** AAGTCACTGCAGCGGCC | ||
<br> | <br> | ||
| - | == | + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-15</p>=== |
| - | |||
| - | <p style="background-color: rgb(238, 221, 130); font-weight: bold;"> | + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">cell culture </p> |
| + | <br> | ||
| + | <b>Investigator:</b>Kerstin<br> | ||
| + | <br> | ||
| + | <b>Topic: </b>cell-culture <br> | ||
| + | <br> | ||
| + | * passaging of CHO w Zeocin | ||
| + | * passaging of HT1080 | ||
| + | * passaging of HEK AAV 293 | ||
| + | * passaging of HeLa | ||
| - | |||
| - | < | + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-17</p>=== |
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">cell culture </p> | ||
| + | <br> | ||
| + | <b>Investigator:</b>Kerstin<br> | ||
| + | <br> | ||
| + | <b>Topic: </b>cell-culture <br> | ||
| + | <br> | ||
| + | * passaging of CHO w Zeocin | ||
| + | * passaging of HT1080 | ||
| + | * passaging of HEK AAV 293 | ||
| + | * passaging of HeLa | ||
| + | * passaging of stably transfected Clones 4.1, 4.2, 4.3, 4,4, 29.1, 29.2, 29.3 with 550µg/ml Hygromycin | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-19</p>=== | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">cell culture </p> | ||
| + | <br> | ||
| + | <b>Investigator:</b>Stefan/Kerstin<br> | ||
| + | <br> | ||
| + | <b>Topic: </b>cell-culture <br> | ||
| + | <br> | ||
| + | * passaging of CHO w Zeocin | ||
| + | * passaging of HT1080 | ||
| + | * passaging of HEK AAV 293 | ||
| + | * passaging of HeLa | ||
| + | |||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">PCR: amplification of EYFP and mcherry with overhang of new RAGE-TMD</p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b> | ||
| + | * Template: EYFP_Bba_E0030 and nano body-geneart-construct | ||
| + | * Phusion-polymerase | ||
| + | * 10x Phusion buffer HF<br> | ||
| + | * dNTPs (10mM) | ||
| + | * eYFP: fwp:nterm. of yfp to TMD; primerVI: c-terminus of eyfp w | ||
| + | * mcherry: rage-fwp:mcherry/rageTMD; rage:endprimer in mcherry/loxp | ||
| + | * Thermocycler<br> | ||
| + | <b>Methods:</b> | ||
| + | '''2x master mix of each: eYFP and mcherry''' | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td>'''reagent''' </td> | ||
| + | <td>'''volume [µL]''' </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10x Phusion HF buffer</td> | ||
| + | <td>20</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>dNTPs</td> | ||
| + | <td>2</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> fwp:nterm. of yfp to TMD; rage-fwp:mcherry/rageTMD </td> | ||
| + | <td>5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> primerVI: c-terminus of eyfp w; rage:endprimer in mcherry/loxp </td> | ||
| + | <td>5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> template EYFP (10ng/µl); template nanobody-geneart-construct (10ng/µl)</td> | ||
| + | <td> 2</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> Phusion Polymerase </td> | ||
| + | <td> 1,0 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> water </td> | ||
| + | <td> 66</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | '''Program''' | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td>'''step''' </td> | ||
| + | <td>'''Temperature [°C]''' </td> | ||
| + | <td>'''duration [s]''' </td> | ||
| + | <td>'''cycles''' </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> initial denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td>30</td> | ||
| + | <td>1 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td> 8 </td> | ||
| + | <td> 30</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> annealing </td> | ||
| + | <td> gradient: 64,7°C/71°C for eYFP; 62,0°C/66,1°C for nanobody-geneart-construct </td> | ||
| + | <td> 10 </td> | ||
| + | <td> 30 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 15 </td> | ||
| + | <td> 30</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> final elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 600 </td> | ||
| + | <td> 1 </td> | ||
| + | </tr> | ||
| + | <tr><td> cooling </td> | ||
| + | <td> 8 </td> | ||
| + | <td> ∞ </td> | ||
| + | <td> 1 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;">Topic: preparative gelelectrophoresis of amplified EYFP and mcherry with RAGE-TMD and RAGE-signalpeptide overhangs</p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
<b>Materials:</b><br> | <b>Materials:</b><br> | ||
| + | * agarose | ||
| + | * 1xTAE-buffer | ||
| + | * 10xFD Green Buffer <br> | ||
| + | <b>Method:</b><br> | ||
| + | * 1% agarosegel, 55ml | ||
| + | * 120V | ||
| + | <b>Results:</b><br> | ||
| + | [[File:UP12_2012-10-20-preprgel_eyfp_mcherry.jpg|350px]] | ||
| + | * amplified eYFP and mcherry showed sufficient bp-size after PCR<br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * gel extraction and concentration measurement | ||
| + | * amplification of scFv-fragment and nanobody/Fc | ||
| - | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> gel extraction of amplified eYFP and mcherry out of 1% agarosegel </p> | ||
| + | <b>Investigators:</b>Sascha<br> | ||
| + | <b>Results:</b> | ||
| + | <br> | ||
| + | [eYFP] = <br> | ||
| + | [mcherry] = <br> | ||
| + | <b>Further Taks:</b> | ||
| + | * extension PCR of scFv and nanobody | ||
| + | <br> | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);"> 2012-10-20</p>=== | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">PCR: amplification of nanobody/Fc with overhang of new RAGE-TMD</p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b> | ||
| + | * Template: nanobody-geneart-construct | ||
| + | * Phusion-polymerase | ||
| + | * 10x Phusion buffer GC<br> | ||
| + | * dNTPs (10mM) | ||
| + | * nanobody-geneart-construct: rage:startprimer-fw_nano/Fc; rage-rv:Fc to rageTMD | ||
| + | * Thermocycler<br> | ||
| + | <b>Methods:</b> | ||
| + | '''2x master mix of each: eYFP and mcherry''' | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td>'''reagent''' </td> | ||
| + | <td>'''volume [µL]''' </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10x Phusion BC buffer</td> | ||
| + | <td>20</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>dNTPs</td> | ||
| + | <td>2</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> rage:startprimer-fw_nano/Fc </td> | ||
| + | <td>5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> rage-rv:Fc to rageTMD </td> | ||
| + | <td>5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> template nanobody-geneart-construct (10ng/µl)</td> | ||
| + | <td> 6</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> Phusion Polymerase </td> | ||
| + | <td> 1,0 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> water </td> | ||
| + | <td> 92</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | '''Program''' | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td>'''step''' </td> | ||
| + | <td>'''Temperature [°C]''' </td> | ||
| + | <td>'''duration [s]''' </td> | ||
| + | <td>'''cycles''' </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> initial denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td>30</td> | ||
| + | <td>1 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td> 8 </td> | ||
| + | <td> 30</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> annealing </td> | ||
| + | <td> 520°C/62°C/65,5°C </td> | ||
| + | <td> 10 </td> | ||
| + | <td> 30 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 15 </td> | ||
| + | <td> 30</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> final elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 600 </td> | ||
| + | <td> 1 </td> | ||
| + | </tr> | ||
| + | <tr><td> cooling </td> | ||
| + | <td> 8 </td> | ||
| + | <td> ∞ </td> | ||
| + | <td> 1 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;">Topic: preparative gelelectrophoresis of amplified nanobody/Fc with RAGE-TMD overhangs</p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * agarose | ||
| + | * 1xTAE-buffer | ||
| + | * 10xFD Green Buffer <br> | ||
<b>Method:</b><br> | <b>Method:</b><br> | ||
| + | * 1% agarosegel, 55ml | ||
| + | * 120V | ||
| + | <b>Results:</b><br> | ||
| + | [[File:UP12_2012-10-20-nanoassemlby.jpg|300px]] | ||
| + | * amplified nanobody/Fc showed sufficient bp-size @ 1252bp <br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * gel extraction and concentrations measurement | ||
| - | + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> gel extraction of amplified eYFP and mcherry out of 1% agarosegel </p> | |
| + | <b>Investigators:</b>Sascha<br> | ||
| + | <b>Results:</b> | ||
| + | [nanobody/FC] = 2,5ng/µl</b> | ||
| + | <b>Further Taks:</b> | ||
| + | * extension PCR of scFv and nanobody | ||
| + | * assembly-PCR of nanobody/Fc with mherry via RAGE-transmembrane domain | ||
| + | |||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">PCR: assembly-PCR of nanobody/Fc with mcherry over RAGE-TMD</p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b> | ||
| + | * Template: nanobody/FC and mcherry with TMD-overhangs | ||
| + | * Phusion-polymerase | ||
| + | * 10x Phusion buffer GC<br> | ||
| + | * dNTPs (10mM) | ||
| + | * Thermocycler<br> | ||
| + | <b>Methods:</b> | ||
| + | '''2x master mix of each: eYFP and mcherry''' | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td>'''reagent''' </td> | ||
| + | <td>'''volume [µL]''' </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10x Phusion BC buffer</td> | ||
| + | <td>20</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>dNTPs</td> | ||
| + | <td>2</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> rage:startprimer-fw_nano/Fc was added after 15 assembling cycles</td> | ||
| + | <td>5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> rage:endprimer in mcherry/loxp was added after 15 assembling cycles</td> | ||
| + | <td>5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> templates: nanobody/FC with TMD-overhangs ; mcherry with TMD-overhangs (10ng/µl)</td> | ||
| + | <td> 2µl of mcherry and 8µl of nanobody/Fc </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> Phusion Polymerase </td> | ||
| + | <td> 1,0 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> water </td> | ||
| + | <td> 57</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | '''Program''' | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td>'''step''' </td> | ||
| + | <td>'''Temperature [°C]''' </td> | ||
| + | <td>'''duration [s]''' </td> | ||
| + | <td>'''cycles''' </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> initial denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td>30</td> | ||
| + | <td>15 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td> 10 </td> | ||
| + | <td> 15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> annealing </td> | ||
| + | <td> 60°C/70°C </td> | ||
| + | <td> 12</td> | ||
| + | <td> 15 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 38 </td> | ||
| + | <td> 15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> initial denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td>30</td> | ||
| + | <td>15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td> 10 </td> | ||
| + | <td> 15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> annealing </td> | ||
| + | <td> 60°C/70°C </td> | ||
| + | <td> 12</td> | ||
| + | <td> 15 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 38 </td> | ||
| + | <td> 15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> final elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 600 </td> | ||
| + | <td> 1 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> cooling </td> | ||
| + | <td> 8 </td> | ||
| + | <td> ∞ </td> | ||
| + | <td> 1 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;">Topic: analytical gelelectrophoresis of amplified assembled nanobody/Fc with RAGE-TMD overhangs</p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * agarose | ||
| + | * 1xTAE-buffer | ||
| + | * 10xFD Green Buffer <br> | ||
| + | <b>Method:</b><br> | ||
| + | * 1% agarosegel, 55ml | ||
| + | * 120V | ||
<b>Results:</b><br> | <b>Results:</b><br> | ||
| + | [[File:UP12_2012-10-21-amplified_assembled nano.jpg|100px ]] | ||
| + | * assembled nanobody/Fc-RAGE-TMD-mcherry showed sufficient bp-size: 2,1kb<br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * PCR-Clean up of 60°C-assembling PCR-mix | ||
| + | * amplification of assembeled nanobody/Fc-RAGE-TMD-mcherry | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> PCR-Clean up of preparative gel electrophoresis of assembled nanobody/Fc-RAGE-TMD-mcherry and concentration measurement</p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) <br> | ||
| + | <b>Methods:</b> | ||
| + | * according to CleanUp-protocol | ||
| + | <b>Results:</b> | ||
| + | [assembled nanobody/Fc-RAGE-TMD-mcherry ] = <br> | ||
| + | <b>Further Taks:</b> | ||
| + | * preparative gel electrophoresis of assembled nanobody/Fc-RAGE-TMD-mcherry | ||
| + | |||
| + | <br> | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);"> 2012-10-21</p>=== | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;">Topic: preaparative gel electrophoresis of assembled nanobody/Fc-RAGE-TMD-mcherry </p> | ||
| + | <b>Investigator:</b> Stefan<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * agarose | ||
| + | * 1xTAE-buffer | ||
| + | * 10xFD Green Buffer <br> | ||
| + | <b>Method:</b><br> | ||
| + | * 1% agarosegel, 55ml | ||
| + | * 120V | ||
| + | <b>Results:</b><br> | ||
| + | * assembled nanobody/Fc-RAGE-TMD-mcherry showed sufficient bp-size: 2,1kb<br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * gel extraction | ||
| + | * cloning into pcDNA5FRT | ||
| + | |||
| + | <br> | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-22</p>=== | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">cell culture </p> | ||
| + | <br> | ||
| + | <b>Investigator:</b>Kerstin<br> | ||
| + | <br> | ||
| + | <b>Topic: </b>cell-culture <br> | ||
| + | <br> | ||
| + | * passaging of CHO w Zeocin | ||
| + | * passaging of HT1080 | ||
| + | * passaging of HEK AAV 293 | ||
| + | * passaging of HeLa | ||
| + | * transfection of stably transfected clone 29.1 with Cre Recombinase | ||
| + | * seeding of HT1080 in Ibidi Dish for infection with virus | ||
| + | |||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);"> 2012-10-22</p>=== | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> gel extraction of assembled nanobody/Fc-RAGE-TMD-mcherry out of 1% agarosegel </p> | ||
| + | <b>Investigators:</b>Sascha<br> | ||
| + | <b>Results:</b> | ||
| + | [assembled/amplified nanobody/Fc-RAGE-TMD-mcherry] = 60,4ng/µl<br> | ||
| + | <b>Further Taks:</b> | ||
| + | * cloning into pcDNA5FRT | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> preparative digestion of assembled/amplified nanobody/Fc-RAGE-TMD-mcherry with NheI and ApaI </p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * Fast Digest NheI | ||
| + | * Fast Digest ApaI | ||
| + | * 10x FD Green Buffer | ||
| + | * assembled/amplified nanobody/Fc-RAGE-TMD-mcherry | ||
| + | * sterile water | ||
| + | <br> | ||
| + | <b>Method:</b><br> | ||
| + | 2x<br> | ||
| + | * 20µl pcdna5frt (60,4ng/µl) | ||
| + | * 2µl NheI | ||
| + | * 2µl ApaI | ||
| + | * 3µl 10x FD Green Buffer | ||
| + | * 4µl sterile water | ||
| + | * digestion for 1h at 37°C<br> | ||
| + | <b>>Results:</b><br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * PCR-Clean Up | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> PCR-Clean up of digested assembled/amplified nanobody/Fc-RAGE-TMD-mcherry and concentration measurement</p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel) <br> | ||
| + | <b>Methods:</b> | ||
| + | * according to CleanUp-protocol | ||
| + | <b>Results:</b> | ||
| + | [assembled/amplified nanobody/Fc-RAGE-TMD-mcherry] = 6ng/µl<br> | ||
| + | <b>Further Taks:</b> | ||
| + | * ligation with pcDNA5FRT (dephos +dig) | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> ligation of digested dephosporylated pcDNA5FRT (NheI+ApaI) and digested assembled/amplified nanobody/Fc-RAGE-TMD-mcherry (NheI+ApaI) </p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * ligation calculator: http://www.insilico.uni-duesseldorf.de/Lig_Input.html | ||
| + | * T4 DNA ligase | ||
| + | * ligase buffer | ||
| + | * digested geneart-nanobody-construct | ||
| + | * digested and dephosporylated pcDNA5FRT | ||
| + | <br> | ||
| + | <b>Method: ligation-ratio--> 1:3</b><br>: | ||
| + | * 1µl T4 DNA ligase | ||
| + | * 2µl T4 DNA-ligase buffer | ||
| + | * 15 ng dig. pcDNA5FRT | ||
| + | * 15 ng insert (assembled/amplified nanobody/Fc-RAGE-TMD-mcherry) | ||
| + | * sterile water ad 20µl<br> | ||
| + | |||
| + | * 1:0 religation; same mix without insert | ||
| + | |||
| + | <b>Further Tasks:</b><br> | ||
| + | * transformation of ligation mix into XL1-blue competent <i>E. coli</i> cells | ||
| + | <br> | ||
| + | |||
| + | * incubation for 60min at RT | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * transformation of ligation mix into XL1-blue competent <i>E. coli</i> cells | ||
| + | <br> | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> Transformation of ligated pcDNA5FRT- assembled/amplified nanobody/Fc-RAGE-TMD-mcherry into new XL1-blue competent <i>E. coli</i> cells</p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * Bunsen Burner, Agar Plate with Ampicillin, 37 °C thermo mixer, centrifuge,<br> | ||
| + | * 12 µl of each ligation mix | ||
| + | * icebox | ||
| + | * new competent <i>E. coli</i> cells (XL 1) | ||
| + | <br> | ||
| + | <b>Method:</b><br> | ||
| + | * according to manual | ||
| + | * 20µl of resuspended cell-suspension were plated on a LB-Amp-plate | ||
| + | * incubation o.n. at 37°C | ||
| + | <br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * picking clones | ||
| + | <br> | ||
| + | |||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">PCR: amplification of scFv with overhang of new RAGE-TMD</p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b> | ||
| + | * Template: scFv_Bba pks…bla | ||
| + | * Phusion-polymerase | ||
| + | * 10x Phusion buffer HF<br> | ||
| + | * dNTPs (10mM) | ||
| + | * rvp:scfv+overhang to TEV/TMD; primerI-fw:rage-signal, xbaI, | ||
| + | * Thermocycler<br> | ||
| + | <b>Methods:</b> | ||
| + | '''3x master mix of each: scFv''' | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td>'''reagent''' </td> | ||
| + | <td>'''volume [µL]''' </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10x Phusion HF buffer</td> | ||
| + | <td>30</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>dNTPs</td> | ||
| + | <td>3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> scfv+overhang to TEV/TMD </td> | ||
| + | <td>5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> primerI-fw:rage-signal, xbaI </td> | ||
| + | <td>5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> template scFv_Bba pks…bla (10ng/µl)</td> | ||
| + | <td> 3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> Phusion Polymerase </td> | ||
| + | <td> 1,5 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> water </td> | ||
| + | <td>195</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | '''Program''' | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td>'''step''' </td> | ||
| + | <td>'''Temperature [°C]''' </td> | ||
| + | <td>'''duration [s]''' </td> | ||
| + | <td>'''cycles''' </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> initial denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td>30</td> | ||
| + | <td>1 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td> 8 </td> | ||
| + | <td> 30</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> annealing </td> | ||
| + | <td> 53°C/60°C/71°C </td> | ||
| + | <td> 10 </td> | ||
| + | <td> 30 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 15 </td> | ||
| + | <td> 30</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> final elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 600 </td> | ||
| + | <td> 1 </td> | ||
| + | </tr> | ||
| + | <tr><td> cooling </td> | ||
| + | <td> 8 </td> | ||
| + | <td> ∞ </td> | ||
| + | <td> 1 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;">Topic: preparative gel electrophoresis of amplified scFv with RAGE-TMD overhangs</p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * agarose | ||
| + | * 1xTAE-buffer | ||
| + | * 10xFD Green Buffer <br> | ||
| + | <b>Method:</b><br> | ||
| + | * 1% agarosegel, 55ml | ||
| + | * 120V | ||
| + | <b>Results:</b><br> | ||
| + | [[File:UP12_2012-10-22-scFVRAGEoverhang.jpg|300px ]] | ||
| + | * amplified scFv showed sufficient bp-size @ 9102bp <br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * PCR-Clean Up of amplified scFv | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> gel extraction of amplified scFv-RAGE-TMD out of 1% agarosegel </p> | ||
| + | <b>Investigators:</b>Sascha<br> | ||
| + | <b>Results:</b> | ||
| + | [amplified scFv-RAGE-TMD] = 60,4ng/µl<br> | ||
| + | <b>Further Taks:</b> | ||
| + | * assembling with eYFP-RAGE overhang | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">PCR: assembly-PCR of scFv with eYFP over RAGE-TMD</p> | ||
| + | <b>Investigators:</b> Sascha<br> | ||
| + | <b>Materials:</b> | ||
| + | * Template: scFv with eYFP with TMD-overhangs | ||
| + | * Phusion-polymerase | ||
| + | * 10x Phusion buffer GC<br> | ||
| + | * dNTPs (10mM) | ||
| + | * Thermocycler<br> | ||
| + | <b>Methods:</b> | ||
| + | '''3x master mix of each: eYFP ''' | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td>'''reagent''' </td> | ||
| + | <td>'''volume [µL]''' </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10x Phusion GC buffer</td> | ||
| + | <td>30</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>dNTPs</td> | ||
| + | <td>3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> scFv:startprimer-fw, was added after 15 assembling cycles</td> | ||
| + | <td>7,5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> scFv:endprimer-rv was added after 15 assembling cycles</td> | ||
| + | <td>7,5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> templates: scFv with TMD-overhangs and RAGTE-signalpeptide (10ng/µl); eYFP with TMD-overhangs (10ng/µl)</td> | ||
| + | <td> 1µl of eYFP and 1µl of scFv </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> Phusion Polymerase </td> | ||
| + | <td> 1,5 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> water </td> | ||
| + | <td> 96,5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | '''Program''' | ||
| + | <table border=1> | ||
| + | <tr> | ||
| + | <td>'''step''' </td> | ||
| + | <td>'''Temperature [°C]''' </td> | ||
| + | <td>'''duration [s]''' </td> | ||
| + | <td>'''cycles''' </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> initial denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td>30</td> | ||
| + | <td>15 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td> 10 </td> | ||
| + | <td> 15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> annealing </td> | ||
| + | <td> 60°C/70°C </td> | ||
| + | <td> 12</td> | ||
| + | <td> 15 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 38 </td> | ||
| + | <td> 15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> initial denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td>30</td> | ||
| + | <td>15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> denaturation </td> | ||
| + | <td> 98 </td> | ||
| + | <td> 10 </td> | ||
| + | <td> 15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> annealing </td> | ||
| + | <td> 60°C/70°C </td> | ||
| + | <td> 12</td> | ||
| + | <td> 15 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 38 </td> | ||
| + | <td> 15</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> final elongation </td> | ||
| + | <td> 72 </td> | ||
| + | <td> 600 </td> | ||
| + | <td> 1 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> cooling </td> | ||
| + | <td> 8 </td> | ||
| + | <td> ∞ </td> | ||
| + | <td> 1 </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | </table> | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;">Topic: analytical gelelectrophoresis of assembled scFv-RAGE-eYFP construct</p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * agarose | ||
| + | * 1xTAE-buffer | ||
| + | * 10xFD Green Buffer <br> | ||
| + | <b>Method:</b><br> | ||
| + | * 1% agarosegel, 55ml | ||
| + | * 120V | ||
| + | <b>Results:</b><br> | ||
| + | [[File:UP12_2012-10-23-assembled RAGEscfv.jpg|300px]] | ||
| + | * assembled scFv-RAGE-eYFP construct showed sufficient bp-size: 1,7kb<br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * PCR-Clean up of 60°C-assembling PCR-mix | ||
| + | * amplification of assembeled scFv-RAGE-eYFP construct | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-23</p>=== | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">cell culture </p> | ||
| + | <br> | ||
| + | <b>Investigator:</b>Stefan<br> | ||
| + | <br> | ||
| + | <b>Topic: </b>cell-culture <br> | ||
| + | <br> | ||
| + | * seeding of CHO and HeLa in Ibidi Dishes for transfection with new Nanobody RAGE construct | ||
| + | * infection with Virus (CFP on surface and YFP as GOI, AAV with Sortase motif) | ||
| + | <br> | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> gelextracton of scFv-RAGE-TMD-EYFP in Flp-In vector construct out of 1% agarosegel </p> | ||
| + | <b>Investigators:</b>Maria<br> | ||
| + | <b>Aim:</b> gelextraction and preparation of cleaned scFv-RAGE-TMD construct <br> | ||
| + | <b>Materials:</b><br> | ||
| + | * Gel-Clean-Up Kit | ||
| + | <br> | ||
| + | <b>Method:</b><br> | ||
| + | * according to manual | ||
| + | <br> | ||
| + | <b>Results:</b><br> | ||
| + | * 32,6 ng/µl | ||
| + | <br> | ||
| + | <b>Further tasks:</b><br> | ||
| + | * digestion with NheI and ApaI | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> preparative digestion of scFv-RAGE-TMD construct with NheI and ApaI </p> | ||
| + | <b>Investigator:</b> Maria<br> | ||
| + | <b>Aim:</b> digestion of scFv-RAGE-TMD construct with NheI and ApaI for ligation into Flp-in vector <br> | ||
| + | <b>Materials:</b><br> | ||
| + | * Fast Digest NheI | ||
| + | * Fast Digest ApaI | ||
| + | * 10x FD Green Buffer | ||
| + | * scFv construct | ||
| + | * sterile water | ||
| + | <br> | ||
| + | <b>Method:</b><br> | ||
| + | * 18,4µl scFv construct | ||
| + | * 2µl NheI | ||
| + | * 2µl ApaI | ||
| + | * 3µl 10x FD Green Buffer | ||
| + | * 4,6µl sterile water | ||
| + | * digestion for 2,5h at 37°C | ||
| + | <br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * PCR clean-up | ||
| + | * ligation into | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> PCR clean-up of scFv-RAGE-TMD construct </p> | ||
| + | <b>Investigator:</b>Maria<br> | ||
| + | <b>Aim:</b> cleaning of scFv-RAGE-TMD<br> | ||
| + | <b>Materials:</b> | ||
| + | * PCR-Clean-Up Kit | ||
| + | <b>Method:</b> | ||
| + | * according to manual | ||
| + | <b>Results:</b> | ||
| + | * concentration of cleaned scFv-RAGE-TMD = 19,7 ng/µl | ||
| + | <br/> | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> ligation of digested scFv-RAGE-TMD construct and dephosporylated Flp-In vector </p> | ||
| + | <b>Investigator:</b> Maria<br> | ||
| + | <b>Aim:</b> ligation of digested scFv construct with Flp-In vector<br> | ||
| + | <br> | ||
| + | <b>Materials:</b><br> | ||
| + | * ligation calculator: http://www.insilico.uni-duesseldorf.de/Lig_Input.html | ||
| + | * T4 DNA ligae | ||
| + | * ligase buffer | ||
| + | * digested scFv construct | ||
| + | * digested and dephosporylated Flp-In vector | ||
| + | <br> | ||
| + | <b>Method: ligation-ratio--> 1:3</b><br> | ||
| + | * 1µl T4 DNA ligase | ||
| + | * 2,5µl T4 DNA-ligase buffer | ||
| + | * 3,3µl digested scFv (19,7ng/µl) | ||
| + | * 1µl digested Flp-In vector (60,4ng/µl) | ||
| + | * 12,2µl sterile water | ||
| + | <br> | ||
| + | * incubation for 1h at RT | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * transformation of ligation mix into XL1-blue competent <i>E. coli</i> cells | ||
| + | <br> | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;"> Transformation of ligated scFv-Flp-In vector into new XL1-blue competent <i>E. coli</i> cells</p> | ||
| + | <b>Investigator:</b> Sascha<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * Bunsen Burner, Agar Plate with Ampicillin, 37 °C thermo mixer, centrifuge,<br> | ||
| + | * 10 µl of cFv-Flp-In vector | ||
| + | * icebox | ||
| + | * competent <i>E. coli</i> cells (XL 1) | ||
| + | <br> | ||
| + | <b>Method:</b><br> | ||
| + | * according to manual | ||
| + | * 20µl of resuspended cell-suspension were plated on a LB-Cm-plate | ||
| + | * incubation o.n. at 37°C | ||
| + | <br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * picking clones | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-24</p>=== | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">cell culture </p> | ||
| + | <br> | ||
| + | <b>Investigator:</b>Stefan/Kerstin<br> | ||
| + | <br> | ||
| + | <b>Topic: </b>cell-culture <br> | ||
| + | <br> | ||
| + | * Transfection of new Nanobody RAGE construct in CHO and HeLa | ||
| + | * seeding of CHO and HeLa in Ibidi Dishes for transfection with new scFv RAGE construct | ||
| + | * passaging of CHO w Zeocin | ||
| + | * passaging of HT1080 | ||
| + | * passaging of HEK AAV 293 | ||
| + | * passaging of HeLa | ||
| + | <br> | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">Endotoxin free preparation of nanobody/Fc-RAGE-TMD-mcherry</p> | ||
| + | <br> | ||
| + | <b>Investigator:</b>Maria<br> | ||
| + | <br> | ||
| + | <b>Materials: </b>endotoxin free Mediprep kit, overnight culture<br> | ||
| + | <br> | ||
| + | <b>Methods: </b>according to manual <br> | ||
| + | <br> | ||
| + | <b>Results: </b><b/> | ||
| + | clone 2: 880,3 ng/µl<br> | ||
| + | <br> | ||
| + | <b>Further tasks: </b>transient and stable transfection of CHO cells<br> | ||
| + | <br> | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-25</p>=== | ||
| + | |||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">cell culture </p> | ||
| + | <br> | ||
| + | <b>Investigator:</b>Stefan/Kerstin<br> | ||
| + | <br> | ||
| + | <b>Topic: </b>cell-culture <br> | ||
| + | <br> | ||
| + | * transfection of new scFv RAGE construct | ||
| + | * transfection of TAL-AID from cooperation with Freiburg iGEM Team in CHO cells (co-transfection with clone 4) | ||
| + | * purification of Nanobody from supernatant of Cre recombinase transfected cells with magnetic beads | ||
| + | * Western Blot of purified Nanobody | ||
| + | <br> | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;">Mini Prep of scFv-Flp-In vector clones </p> | ||
| + | <br> | ||
| + | <b>Investigator:</b>Maria<br> | ||
| + | <br> | ||
| + | <b>Materials: </b>overnight cultures scFv-Flp-In clones, Mini prep kit <br> | ||
| + | <br> | ||
| + | <b>Methods: </b>according to manual <br> | ||
| + | <br> | ||
| + | <b>Results: </b> | ||
| + | Clon I: 514,8 ng/µl<br> | ||
| + | Clon II: 673 ng/µl<br> | ||
| + | Clon III: 639,7 ng/µl<br> | ||
| + | Clon VI: 499 ng/µl<br> | ||
| + | <br> | ||
| + | <b>Further tasks: </b>analytical digestion, analytical gelelectrophoresis<br> | ||
| + | <br> | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">analytical digestion of ligated scFv-Flp-In construct</p> | ||
| + | <b>Investigators:</b>Maria<br> | ||
| + | <br> | ||
| + | <b>Materials:</b> | ||
| + | * all 4 prepared scFv-Flp-In clones | ||
| + | * FastDigest StuI, SpeI and PstI | ||
| + | * 10x FD Green Buffer | ||
| + | * sterile water | ||
| + | <b>Method:</b> | ||
| + | I:<br> | ||
| + | * 10µl mix: 1µl StuI, 1µl 10x FD Green Buffer, 1µl of each clone respectively (approximately 250 ng DNA), sterile water add to 10µl | ||
| + | II:<br> | ||
| + | * 10µl mix: 1µl StuI, 1µl 10x FD Green Buffer, 1µl of each clone respectively (approximately 250 ng DNA), sterile water add to 10µl | ||
| + | * incubation at 37°C for 30 min | ||
| + | <b>further tasks:</b> | ||
| + | * analytical gelelectrophoresis | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;">Gelelectrophoresis of analytical digested scFv-Flp-In construct </p> | ||
| + | <b>Investigators:</b>Maria<br> | ||
| + | |||
| + | <b>Aim:</b> checking plasmid-size after ligation of Flp-In vector with scFv-RAGE-TMD in 1% agarosegel<br> | ||
| + | <b>Materials:</b><br> | ||
| + | * agarose | ||
| + | * 1xTAE-buffer | ||
| + | * 10xFD Green Buffer | ||
| + | <br> | ||
| + | <b>Method:</b><br> | ||
| + | * 1% agarosegel, 100ml | ||
| + | * 120 V | ||
| + | <b>Results:</b><br> | ||
| + | * ligation successful <br> | ||
| + | <b>Further Tasks:</b><br> | ||
| + | * endotoxin free of clone | ||
| + | <p style="background-color: rgb(238, 221, 130); font-weight:bold;">Endotoxin free preparation of scFv-RAGE-TMD</p> | ||
| + | <br> | ||
| + | <b>Investigator:</b>Maria<br> | ||
| + | <br> | ||
| + | <b>Materials: </b>endotoxin free Mediprep kit, overnight culture<br> | ||
| + | <br> | ||
| + | <b>Methods: </b>according to manual <br> | ||
| + | <br> | ||
| + | <b>Results: </b><b/> | ||
| + | clone 1: 2349,7 ng/µl<br> | ||
| + | <br> | ||
| + | <b>Further tasks: </b>transient and stable transfection of CHO cells<br> | ||
| + | <br> | ||
| + | |||
| + | ==Virus== | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-19</p>=== | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;"> EGFRd3 purification</p> | ||
| + | |||
| + | <b>Investigator: </b> Tobias/Xenia<br> | ||
| + | |||
| + | <b> Aim: </b> Periplasma-Extract<br> | ||
| + | |||
| + | <b>Materials:</b><br> | ||
| + | |||
| + | *<i>E. coli</i> with expressed EGFR | ||
| + | *Extraction buffer (50 mM Tris, 150 mM NaCl, 500 mM sucrose) | ||
| + | *loading buffer (50 mM Tris, 150 mM NaCl, 30 mM imidazol) | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | |||
| + | *resuspend <i>E. coli</i> with extraction buffer | ||
| + | *incubate 2 hours at 4°C | ||
| + | *centrifuge and take supernatant | ||
| + | *dialyze against loading buffer over night | ||
| + | |||
| + | <b>Further tasks:</b> | ||
| + | |||
| + | *purification with Ni-NTA | ||
| + | |||
| + | ===<p style="background-color: rgb(240, 20, 70);">2012-10-20</p>=== | ||
| + | |||
| + | <p style="background-color: rgb(238, 221, 130); font-weight: bold;"> EGFRd3 purification</p> | ||
| + | |||
| + | <b>Investigator: </b> Tobias<br> | ||
| + | |||
| + | <b> Aim: </b> Purification with Ni-NTA<br> | ||
| + | |||
| + | <b>Materials:</b><br> | ||
| + | |||
| + | *protein extract in loading buffer (50 mM Tris, 150 mM NaCl, 30 mM imidazol) | ||
| + | *wash buffer (50 mM Tris, 150 mM NaCl, 30 mM imidazol) | ||
| + | *elution buffer (50 mM Tris, 150 mM NaCl, 250 mM imidazol) | ||
| + | *Ni-NTA column (1 mL volume) | ||
| + | |||
| + | <b>Method:</b><br> | ||
| + | |||
| + | *loading Ni-NTA column with protein extract | ||
| + | *wash column with wash buffer (10fold column volume) | ||
| + | *elution with elution buffer and seperate ca. every 1 ml | ||
| + | *control purity with SDS-PAGE | ||
| + | |||
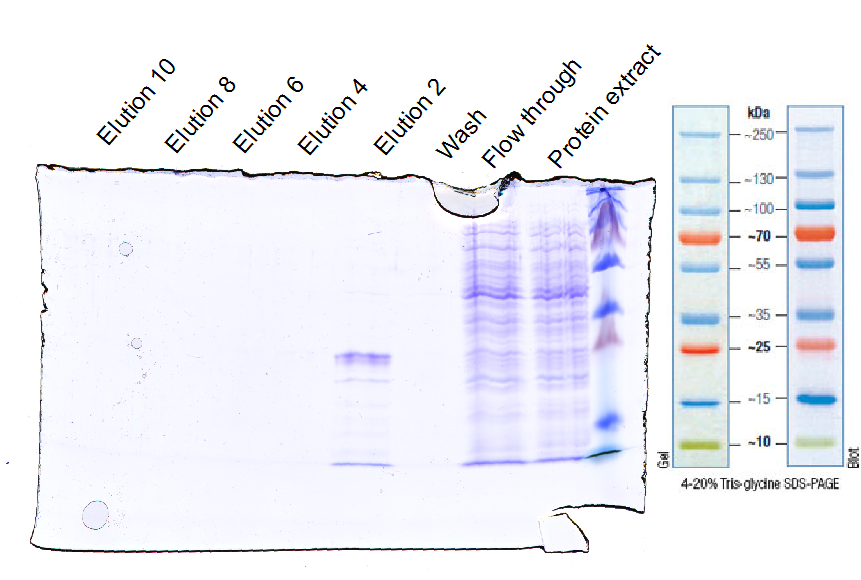
| + | <b>Results:</b> | ||
| - | + | [[File:UP12_SDA-PAGE-2012-10-20.png|500px]] | |
| - | <b>Further tasks</b> | + | <b>Further tasks:</b> |
| - | + | *concentrate purified EGFR and ligate with sortase | |
Latest revision as of 18:24, 26 October 2012
Contents |
AID
2012-10-10
Inoculation of plasmid samples of the 48h retransformation plates (after FACS)
Investigators: Tom
Time: 2012-10-10
Materials:
- LB medium
- Amp stock solution
- plate with cultures:
EGFR-C-AID without NES, with NLS+Kozak sequence+eGFP
(after FACS selection)
Method:
Inoculation of:
5 cultures per plate in 5 ml LB medium + 5µL Amp.
(--> 5 cultures)
Further tasks:
- Miniprep
2012-10-11
Purification of Retrafo plasmids
Investigators: Rico
Results: inoculation was not successful
2012-10-26
Purification of transfected wt AID, modified AID, TAL-AID plasmids cotransfected with small antibody construct and transformation
Investigators: Rico, Stefan, Tom
Method: transfected cells were lysed, plasmids purified and transformed into E. coli cells
Antibody
2012-10-12
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO w Zeocin
- passaging of HT1080
- passaging of HEK AAV 293
- passaging of HeLa
Topic: planning new primer for integration of new RAGE-transmembrane domain and RAGE-signalpeptide
Investigator: Sascha
Materials and Methods: Geneious
Topic: Primer design and ordering for integration of RAGE-transmembrane domain into scFVconstruct and nanobody-geneart construct
Investigator: Sascha
Materials and Methods: Geneious
Results:
- scFv:startprimer-fw
- GCCTCTAGAGCTAGCATGGCAGC
- rvp:scfv+overhang to TEV/TMD
- CGCCCAAGATTCCCAGGGCCAGGGCGAGGGTGCCGCTTCCGCCGCCACCGCTTCCCCCTTGAAAATATAAATTCTCTCCAGATCCCCGTTT
GATCTCCAGTTCTGTCC
- fwp:nterm. of yfp to TMD
- CCCTGGGAATCTTGGGCGGCCTGGGGACCGCTGCCCTCTTGATCGGCGTGATCCTGTGGCAGAGAAGGTCTGGAGTGAGCAAGGGCGAG
GAGC
- scFv:endprimer-rv
- GAGCTGCAGCGGCCG
- rage:startprimer-fw_nano/Fc
- CTTGAATTCGCGGCCG
- rage-rv:Fc to rageTMD
- CGCCCAAGATTCCCAGGGCCAGGGCGAGGGTGCCGCTTCCTAATAACTTCGTATAATGTATGCTATACGAAGTTATCCC
- rage-fwp:mcherry/rageTMD
- CCCTGGGAATCTTGGGCGGCCTGGGGACCGCTGCCCTCTTGATCGGCGTGATCCTGTGGCAGAGAAGGGGCTCCATGGTGTCCAAGG
- rage:endprimer in mcherry/loxp
- AAGTCACTGCAGCGGCC
2012-10-15
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO w Zeocin
- passaging of HT1080
- passaging of HEK AAV 293
- passaging of HeLa
2012-10-17
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO w Zeocin
- passaging of HT1080
- passaging of HEK AAV 293
- passaging of HeLa
- passaging of stably transfected Clones 4.1, 4.2, 4.3, 4,4, 29.1, 29.2, 29.3 with 550µg/ml Hygromycin
2012-10-19
cell culture
Investigator:Stefan/Kerstin
Topic: cell-culture
- passaging of CHO w Zeocin
- passaging of HT1080
- passaging of HEK AAV 293
- passaging of HeLa
PCR: amplification of EYFP and mcherry with overhang of new RAGE-TMD
Investigators: Sascha
Materials:
- Template: EYFP_Bba_E0030 and nano body-geneart-construct
- Phusion-polymerase
- 10x Phusion buffer HF
- dNTPs (10mM)
- eYFP: fwp:nterm. of yfp to TMD; primerVI: c-terminus of eyfp w
- mcherry: rage-fwp:mcherry/rageTMD; rage:endprimer in mcherry/loxp
- Thermocycler
Methods: 2x master mix of each: eYFP and mcherry
| reagent | volume [µL] |
| 10x Phusion HF buffer | 20 |
| dNTPs | 2 |
| fwp:nterm. of yfp to TMD; rage-fwp:mcherry/rageTMD | 5 |
| primerVI: c-terminus of eyfp w; rage:endprimer in mcherry/loxp | 5 |
| template EYFP (10ng/µl); template nanobody-geneart-construct (10ng/µl) | 2 |
| Phusion Polymerase | 1,0 |
| water | 66 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 30 |
| annealing | gradient: 64,7°C/71°C for eYFP; 62,0°C/66,1°C for nanobody-geneart-construct | 10 | 30 |
| elongation | 72 | 15 | 30 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: preparative gelelectrophoresis of amplified EYFP and mcherry with RAGE-TMD and RAGE-signalpeptide overhangs
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 55ml
- 120V
- amplified eYFP and mcherry showed sufficient bp-size after PCR
Further Tasks:
- gel extraction and concentration measurement
- amplification of scFv-fragment and nanobody/Fc
gel extraction of amplified eYFP and mcherry out of 1% agarosegel
Investigators:Sascha
Results:
[eYFP] =
[mcherry] =
Further Taks:
- extension PCR of scFv and nanobody
2012-10-20
PCR: amplification of nanobody/Fc with overhang of new RAGE-TMD
Investigators: Sascha
Materials:
- Template: nanobody-geneart-construct
- Phusion-polymerase
- 10x Phusion buffer GC
- dNTPs (10mM)
- nanobody-geneart-construct: rage:startprimer-fw_nano/Fc; rage-rv:Fc to rageTMD
- Thermocycler
Methods: 2x master mix of each: eYFP and mcherry
| reagent | volume [µL] |
| 10x Phusion BC buffer | 20 |
| dNTPs | 2 |
| rage:startprimer-fw_nano/Fc | 5 |
| rage-rv:Fc to rageTMD | 5 |
| template nanobody-geneart-construct (10ng/µl) | 6 |
| Phusion Polymerase | 1,0 |
| water | 92 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 30 |
| annealing | 520°C/62°C/65,5°C | 10 | 30 |
| elongation | 72 | 15 | 30 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: preparative gelelectrophoresis of amplified nanobody/Fc with RAGE-TMD overhangs
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 55ml
- 120V
- amplified nanobody/Fc showed sufficient bp-size @ 1252bp
Further Tasks:
- gel extraction and concentrations measurement
gel extraction of amplified eYFP and mcherry out of 1% agarosegel
Investigators:Sascha
Results:
[nanobody/FC] = 2,5ng/µl</b>
Further Taks:
- extension PCR of scFv and nanobody
- assembly-PCR of nanobody/Fc with mherry via RAGE-transmembrane domain
PCR: assembly-PCR of nanobody/Fc with mcherry over RAGE-TMD
Investigators: Sascha
Materials:
- Template: nanobody/FC and mcherry with TMD-overhangs
- Phusion-polymerase
- 10x Phusion buffer GC
- dNTPs (10mM)
- Thermocycler
Methods: 2x master mix of each: eYFP and mcherry
| reagent | volume [µL] |
| 10x Phusion BC buffer | 20 |
| dNTPs | 2 |
| rage:startprimer-fw_nano/Fc was added after 15 assembling cycles | 5 |
| rage:endprimer in mcherry/loxp was added after 15 assembling cycles | 5 |
| templates: nanobody/FC with TMD-overhangs ; mcherry with TMD-overhangs (10ng/µl) | 2µl of mcherry and 8µl of nanobody/Fc |
| Phusion Polymerase | 1,0 |
| water | 57 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 15 |
| denaturation | 98 | 10 | 15 |
| annealing | 60°C/70°C | 12 | 15 |
| elongation | 72 | 38 | 15 |
| initial denaturation | 98 | 30 | 15 |
| denaturation | 98 | 10 | 15 |
| annealing | 60°C/70°C | 12 | 15 |
| elongation | 72 | 38 | 15 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: analytical gelelectrophoresis of amplified assembled nanobody/Fc with RAGE-TMD overhangs
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 55ml
- 120V
Results:
100px
- assembled nanobody/Fc-RAGE-TMD-mcherry showed sufficient bp-size: 2,1kb
Further Tasks:
- PCR-Clean up of 60°C-assembling PCR-mix
- amplification of assembeled nanobody/Fc-RAGE-TMD-mcherry
PCR-Clean up of preparative gel electrophoresis of assembled nanobody/Fc-RAGE-TMD-mcherry and concentration measurement
Investigator: Sascha
Materials:
- NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel)
Methods:
- according to CleanUp-protocol
Results:
[assembled nanobody/Fc-RAGE-TMD-mcherry ] =
Further Taks:
- preparative gel electrophoresis of assembled nanobody/Fc-RAGE-TMD-mcherry
2012-10-21
Topic: preaparative gel electrophoresis of assembled nanobody/Fc-RAGE-TMD-mcherry
Investigator: Stefan
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 55ml
- 120V
Results:
- assembled nanobody/Fc-RAGE-TMD-mcherry showed sufficient bp-size: 2,1kb
Further Tasks:
- gel extraction
- cloning into pcDNA5FRT
2012-10-22
cell culture
Investigator:Kerstin
Topic: cell-culture
- passaging of CHO w Zeocin
- passaging of HT1080
- passaging of HEK AAV 293
- passaging of HeLa
- transfection of stably transfected clone 29.1 with Cre Recombinase
- seeding of HT1080 in Ibidi Dish for infection with virus
2012-10-22
gel extraction of assembled nanobody/Fc-RAGE-TMD-mcherry out of 1% agarosegel
Investigators:Sascha
Results:
[assembled/amplified nanobody/Fc-RAGE-TMD-mcherry] = 60,4ng/µl
Further Taks:
- cloning into pcDNA5FRT
preparative digestion of assembled/amplified nanobody/Fc-RAGE-TMD-mcherry with NheI and ApaI
Investigator: Sascha
Materials:
- Fast Digest NheI
- Fast Digest ApaI
- 10x FD Green Buffer
- assembled/amplified nanobody/Fc-RAGE-TMD-mcherry
- sterile water
Method:
2x
- 20µl pcdna5frt (60,4ng/µl)
- 2µl NheI
- 2µl ApaI
- 3µl 10x FD Green Buffer
- 4µl sterile water
- digestion for 1h at 37°C
>Results:
Further Tasks:
- PCR-Clean Up
PCR-Clean up of digested assembled/amplified nanobody/Fc-RAGE-TMD-mcherry and concentration measurement
Investigator: Sascha
Materials:
- NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel)
Methods:
- according to CleanUp-protocol
Results:
[assembled/amplified nanobody/Fc-RAGE-TMD-mcherry] = 6ng/µl
Further Taks:
- ligation with pcDNA5FRT (dephos +dig)
ligation of digested dephosporylated pcDNA5FRT (NheI+ApaI) and digested assembled/amplified nanobody/Fc-RAGE-TMD-mcherry (NheI+ApaI)
Investigator: Sascha
Materials:
- ligation calculator: http://www.insilico.uni-duesseldorf.de/Lig_Input.html
- T4 DNA ligase
- ligase buffer
- digested geneart-nanobody-construct
- digested and dephosporylated pcDNA5FRT
Method: ligation-ratio--> 1:3
:
- 1µl T4 DNA ligase
- 2µl T4 DNA-ligase buffer
- 15 ng dig. pcDNA5FRT
- 15 ng insert (assembled/amplified nanobody/Fc-RAGE-TMD-mcherry)
- sterile water ad 20µl
- 1:0 religation; same mix without insert
Further Tasks:
- transformation of ligation mix into XL1-blue competent E. coli cells
- incubation for 60min at RT
Further Tasks:
- transformation of ligation mix into XL1-blue competent E. coli cells
Transformation of ligated pcDNA5FRT- assembled/amplified nanobody/Fc-RAGE-TMD-mcherry into new XL1-blue competent E. coli cells
Investigator: Sascha
Materials:
- Bunsen Burner, Agar Plate with Ampicillin, 37 °C thermo mixer, centrifuge,
- 12 µl of each ligation mix
- icebox
- new competent E. coli cells (XL 1)
Method:
- according to manual
- 20µl of resuspended cell-suspension were plated on a LB-Amp-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones
PCR: amplification of scFv with overhang of new RAGE-TMD
Investigators: Sascha
Materials:
- Template: scFv_Bba pks…bla
- Phusion-polymerase
- 10x Phusion buffer HF
- dNTPs (10mM)
- rvp:scfv+overhang to TEV/TMD; primerI-fw:rage-signal, xbaI,
- Thermocycler
Methods: 3x master mix of each: scFv
| reagent | volume [µL] |
| 10x Phusion HF buffer | 30 |
| dNTPs | 3 |
| scfv+overhang to TEV/TMD | 5 |
| primerI-fw:rage-signal, xbaI | 5 |
| template scFv_Bba pks…bla (10ng/µl) | 3 |
| Phusion Polymerase | 1,5 |
| water | 195 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 1 |
| denaturation | 98 | 8 | 30 |
| annealing | 53°C/60°C/71°C | 10 | 30 |
| elongation | 72 | 15 | 30 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: preparative gel electrophoresis of amplified scFv with RAGE-TMD overhangs
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 55ml
- 120V
- amplified scFv showed sufficient bp-size @ 9102bp
Further Tasks:
- PCR-Clean Up of amplified scFv
gel extraction of amplified scFv-RAGE-TMD out of 1% agarosegel
Investigators:Sascha
Results:
[amplified scFv-RAGE-TMD] = 60,4ng/µl
Further Taks:
- assembling with eYFP-RAGE overhang
PCR: assembly-PCR of scFv with eYFP over RAGE-TMD
Investigators: Sascha
Materials:
- Template: scFv with eYFP with TMD-overhangs
- Phusion-polymerase
- 10x Phusion buffer GC
- dNTPs (10mM)
- Thermocycler
Methods: 3x master mix of each: eYFP
| reagent | volume [µL] |
| 10x Phusion GC buffer | 30 |
| dNTPs | 3 |
| scFv:startprimer-fw, was added after 15 assembling cycles | 7,5 |
| scFv:endprimer-rv was added after 15 assembling cycles | 7,5 |
| templates: scFv with TMD-overhangs and RAGTE-signalpeptide (10ng/µl); eYFP with TMD-overhangs (10ng/µl) | 1µl of eYFP and 1µl of scFv |
| Phusion Polymerase | 1,5 |
| water | 96,5 |
Program
| step | Temperature [°C] | duration [s] | cycles |
| initial denaturation | 98 | 30 | 15 |
| denaturation | 98 | 10 | 15 |
| annealing | 60°C/70°C | 12 | 15 |
| elongation | 72 | 38 | 15 |
| initial denaturation | 98 | 30 | 15 |
| denaturation | 98 | 10 | 15 |
| annealing | 60°C/70°C | 12 | 15 |
| elongation | 72 | 38 | 15 |
| final elongation | 72 | 600 | 1 |
| cooling | 8 | ∞ | 1 |
Topic: analytical gelelectrophoresis of assembled scFv-RAGE-eYFP construct
Investigator: Sascha
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 55ml
- 120V
- assembled scFv-RAGE-eYFP construct showed sufficient bp-size: 1,7kb
Further Tasks:
- PCR-Clean up of 60°C-assembling PCR-mix
- amplification of assembeled scFv-RAGE-eYFP construct
2012-10-23
cell culture
Investigator:Stefan
Topic: cell-culture
- seeding of CHO and HeLa in Ibidi Dishes for transfection with new Nanobody RAGE construct
- infection with Virus (CFP on surface and YFP as GOI, AAV with Sortase motif)
gelextracton of scFv-RAGE-TMD-EYFP in Flp-In vector construct out of 1% agarosegel
Investigators:Maria
Aim: gelextraction and preparation of cleaned scFv-RAGE-TMD construct
Materials:
- Gel-Clean-Up Kit
Method:
- according to manual
Results:
- 32,6 ng/µl
Further tasks:
- digestion with NheI and ApaI
preparative digestion of scFv-RAGE-TMD construct with NheI and ApaI
Investigator: Maria
Aim: digestion of scFv-RAGE-TMD construct with NheI and ApaI for ligation into Flp-in vector
Materials:
- Fast Digest NheI
- Fast Digest ApaI
- 10x FD Green Buffer
- scFv construct
- sterile water
Method:
- 18,4µl scFv construct
- 2µl NheI
- 2µl ApaI
- 3µl 10x FD Green Buffer
- 4,6µl sterile water
- digestion for 2,5h at 37°C
Further Tasks:
- PCR clean-up
- ligation into
PCR clean-up of scFv-RAGE-TMD construct
Investigator:Maria
Aim: cleaning of scFv-RAGE-TMD
Materials:
- PCR-Clean-Up Kit
Method:
- according to manual
Results:
- concentration of cleaned scFv-RAGE-TMD = 19,7 ng/µl
ligation of digested scFv-RAGE-TMD construct and dephosporylated Flp-In vector
Investigator: Maria
Aim: ligation of digested scFv construct with Flp-In vector
Materials:
- ligation calculator: http://www.insilico.uni-duesseldorf.de/Lig_Input.html
- T4 DNA ligae
- ligase buffer
- digested scFv construct
- digested and dephosporylated Flp-In vector
Method: ligation-ratio--> 1:3
- 1µl T4 DNA ligase
- 2,5µl T4 DNA-ligase buffer
- 3,3µl digested scFv (19,7ng/µl)
- 1µl digested Flp-In vector (60,4ng/µl)
- 12,2µl sterile water
- incubation for 1h at RT
Further Tasks:
- transformation of ligation mix into XL1-blue competent E. coli cells
Transformation of ligated scFv-Flp-In vector into new XL1-blue competent E. coli cells
Investigator: Sascha
Materials:
- Bunsen Burner, Agar Plate with Ampicillin, 37 °C thermo mixer, centrifuge,
- 10 µl of cFv-Flp-In vector
- icebox
- competent E. coli cells (XL 1)
Method:
- according to manual
- 20µl of resuspended cell-suspension were plated on a LB-Cm-plate
- incubation o.n. at 37°C
Further Tasks:
- picking clones
2012-10-24
cell culture
Investigator:Stefan/Kerstin
Topic: cell-culture
- Transfection of new Nanobody RAGE construct in CHO and HeLa
- seeding of CHO and HeLa in Ibidi Dishes for transfection with new scFv RAGE construct
- passaging of CHO w Zeocin
- passaging of HT1080
- passaging of HEK AAV 293
- passaging of HeLa
Endotoxin free preparation of nanobody/Fc-RAGE-TMD-mcherry
Investigator:Maria
Materials: endotoxin free Mediprep kit, overnight culture
Methods: according to manual
Results: <b/>
clone 2: 880,3 ng/µl
Further tasks: transient and stable transfection of CHO cells
2012-10-25
cell culture
Investigator:Stefan/Kerstin
Topic: cell-culture
- transfection of new scFv RAGE construct
- transfection of TAL-AID from cooperation with Freiburg iGEM Team in CHO cells (co-transfection with clone 4)
- purification of Nanobody from supernatant of Cre recombinase transfected cells with magnetic beads
- Western Blot of purified Nanobody
Mini Prep of scFv-Flp-In vector clones
Investigator:Maria
Materials: overnight cultures scFv-Flp-In clones, Mini prep kit
Methods: according to manual
Results:
Clon I: 514,8 ng/µl
Clon II: 673 ng/µl
Clon III: 639,7 ng/µl
Clon VI: 499 ng/µl
Further tasks: analytical digestion, analytical gelelectrophoresis
analytical digestion of ligated scFv-Flp-In construct
Investigators:Maria
Materials:
- all 4 prepared scFv-Flp-In clones
- FastDigest StuI, SpeI and PstI
- 10x FD Green Buffer
- sterile water
Method:
I:
- 10µl mix: 1µl StuI, 1µl 10x FD Green Buffer, 1µl of each clone respectively (approximately 250 ng DNA), sterile water add to 10µl
II:
- 10µl mix: 1µl StuI, 1µl 10x FD Green Buffer, 1µl of each clone respectively (approximately 250 ng DNA), sterile water add to 10µl
- incubation at 37°C for 30 min
further tasks:
- analytical gelelectrophoresis
Gelelectrophoresis of analytical digested scFv-Flp-In construct
Investigators:Maria
Aim: checking plasmid-size after ligation of Flp-In vector with scFv-RAGE-TMD in 1% agarosegel
Materials:
- agarose
- 1xTAE-buffer
- 10xFD Green Buffer
Method:
- 1% agarosegel, 100ml
- 120 V
Results:
- ligation successful
Further Tasks:
- endotoxin free of clone
Endotoxin free preparation of scFv-RAGE-TMD
Investigator:Maria
Materials: endotoxin free Mediprep kit, overnight culture
Methods: according to manual
Results: <b/>
clone 1: 2349,7 ng/µl
Further tasks: transient and stable transfection of CHO cells
Virus
2012-10-19
EGFRd3 purification
Investigator: Tobias/Xenia
Aim: Periplasma-Extract
Materials:
- E. coli with expressed EGFR
- Extraction buffer (50 mM Tris, 150 mM NaCl, 500 mM sucrose)
- loading buffer (50 mM Tris, 150 mM NaCl, 30 mM imidazol)
Method:
- resuspend E. coli with extraction buffer
- incubate 2 hours at 4°C
- centrifuge and take supernatant
- dialyze against loading buffer over night
Further tasks:
- purification with Ni-NTA
2012-10-20
EGFRd3 purification
Investigator: Tobias
Aim: Purification with Ni-NTA
Materials:
- protein extract in loading buffer (50 mM Tris, 150 mM NaCl, 30 mM imidazol)
- wash buffer (50 mM Tris, 150 mM NaCl, 30 mM imidazol)
- elution buffer (50 mM Tris, 150 mM NaCl, 250 mM imidazol)
- Ni-NTA column (1 mL volume)
Method:
- loading Ni-NTA column with protein extract
- wash column with wash buffer (10fold column volume)
- elution with elution buffer and seperate ca. every 1 ml
- control purity with SDS-PAGE
Results:
Further tasks:
- concentrate purified EGFR and ligate with sortase
 "
"