Team:SJTU-BioX-Shanghai/Project/project1.3
From 2012.igem.org
AleAlejandro (Talk | contribs) (→Application) |
AleAlejandro (Talk | contribs) (→RNA Signal) |
||
| (16 intermediate revisions not shown) | |||
| Line 28: | Line 28: | ||
{{Template:12SJTU_part_summary_head}} | {{Template:12SJTU_part_summary_head}} | ||
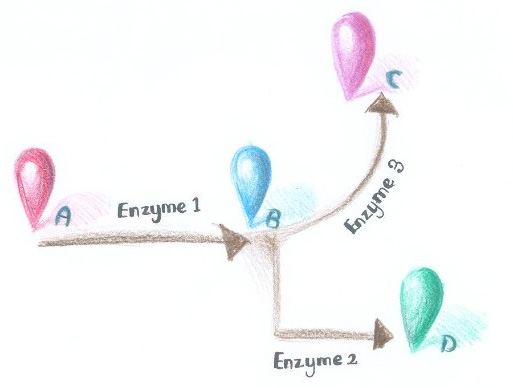
| - | Dynamically and artificially regulating the direction of biochemical pathway ''in vivo'' has remained a challenge for scientists. We were trying to achieve this goal through controlling the aggregation state of different enzymes. If we replace certain constitutively dimerizing proteins in ''Membrane Accelerator'' with signal-induced dimers, then it is possible to dynamically control the direction of branched reactions. | + | Dynamically and artificially regulating the direction of biochemical pathway ''in vivo'' has remained a challenge for scientists. We were trying to achieve this goal through controlling the aggregation state of different enzymes. If we replace certain constitutively dimerizing proteins in ''Membrane Accelerator'' with signal-induced dimers, then it is possible to dynamically control the direction of branched reactions. In this way, a ''Membrane Rudder'' to regulate the direction of biochemical pathway could be created. |
| + | [[Image:12SJTU_divergentreaction.jpg|thumb|400px|right|''Fig.1'' :Demonstration of branched reactions.]] | ||
Here is a simplified example. As is described in Figure 1, when signal that induces the aggregation of Enzyme 1 and 2 is present, the two enzymes would get close to each other, making product D more dominant. On the contrary, when signal that induces the aggregation of Enzyme 1 and 3 is present, Enzyme 1 and 3 get close to each other, so product C is more dominant. | Here is a simplified example. As is described in Figure 1, when signal that induces the aggregation of Enzyme 1 and 2 is present, the two enzymes would get close to each other, making product D more dominant. On the contrary, when signal that induces the aggregation of Enzyme 1 and 3 is present, Enzyme 1 and 3 get close to each other, so product C is more dominant. | ||
There are indeed many signal-induced dimers that commonly exist in nature, which could sense light, chemical, peptide and even RNA signal. | There are indeed many signal-induced dimers that commonly exist in nature, which could sense light, chemical, peptide and even RNA signal. | ||
| - | |||
{{Template:12SJTU_part_summary_foot}} | {{Template:12SJTU_part_summary_foot}} | ||
| Line 55: | Line 55: | ||
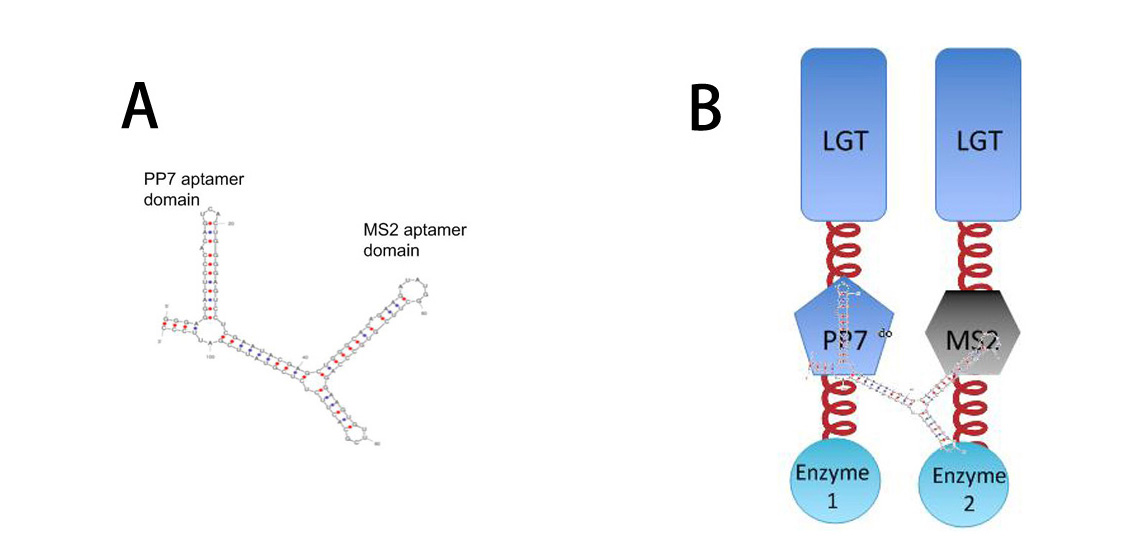
| - | So far, ''Membrane Accelerator'' and ''Membrane Rudder'' are both post- | + | So far, ''Membrane Accelerator'' and ''Membrane Rudder'' are both post-translational controlling device to regulate metabolic flux of the host cell. To connect this relatively isolated post-translational control system to genetic circuits, we employed RNA signal, which is present in cytoplasm. When certain RNA molecule with two aptamer domains is present in cells, their cognate aptamer binding proteins can thus aggregate together. |
| - | [[Image:12SJTU RNASIGNAL.jpg|thumb| | + | [[Image:12SJTU RNASIGNAL.jpg|thumb|650px|center|''Fig.3'' : |
<br> | <br> | ||
'''A''': Sketch of signal RNA molecule (called D0) which consists of PP7 and MS2 aptamer domains. PP7 aptamer binding protein could dimerize with PP7 RNA aptamer domain; MS2 aptamer binding protein could dimerize with MS2 RNA aptamer domain. | '''A''': Sketch of signal RNA molecule (called D0) which consists of PP7 and MS2 aptamer domains. PP7 aptamer binding protein could dimerize with PP7 RNA aptamer domain; MS2 aptamer binding protein could dimerize with MS2 RNA aptamer domain. | ||
| Line 73: | Line 73: | ||
Recruiting those chemical and peptide induced dimers could easily broaden the application field of ''Membrane Rudder''. | Recruiting those chemical and peptide induced dimers could easily broaden the application field of ''Membrane Rudder''. | ||
| + | |||
| + | ==Construction Details== | ||
| + | |||
| + | The construction strategy of ''Membrane Rudder'' is similar to that of ''Membrane Accelerator''. The only difference is that signal induced dimers are introduced in Membrane Anchors so that the aggregation state of Membrane Anchors could be dynamically controlled. | ||
| + | |||
| + | [[File:12SJTU_overview4.gif|center|400px]] | ||
| + | |||
| + | <center>''Fig.4'': Sketch of ''Membrane Rudder''</center> | ||
| + | |||
| + | Through controlling the aggregation state of Membrane Anchor linked with enzymes, we can alter the direction of branched biochemical reactions. | ||
==Application== | ==Application== | ||
| Line 78: | Line 88: | ||
We chose blue light sensor protein -- VVD to test the feasibility of ''Membrane Rudder'' device because of the uniqueness of light signal. | We chose blue light sensor protein -- VVD to test the feasibility of ''Membrane Rudder'' device because of the uniqueness of light signal. | ||
| - | [[Team:SJTU-BioX-Shanghai/Project/project2.1|Violacein and deoxyviolacein synthetic pathway]], a branched enzymatic reaction is recruited to prove that ''Membrane Rudder'' sensing blue light could work as expected. | + | [[Team:SJTU-BioX-Shanghai/Project/project2.1|Violacein and deoxyviolacein synthetic pathway]], a branched enzymatic reaction is recruited to prove that ''Membrane Rudder'' sensing blue light could work as expected. The experiment result confirmed the feasibility of ''Membrane Rudder'' in direction alteration. |
==Reference== | ==Reference== | ||
Latest revision as of 17:01, 26 October 2012
| ||
|
 "
"