Team:LMU-Munich/Spore Coat Proteins/cloning
From 2012.igem.org
Franzi.Duerr (Talk | contribs) |
|||
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<!-- Include the next line at the beginning of every page --> | <!-- Include the next line at the beginning of every page --> | ||
{{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_eppis.resized.jpg|3}} | {{:Team:LMU-Munich/Templates/Page Header|File:Team-LMU_eppis.resized.jpg|3}} | ||
| + | [[File:LMU Glow Spore2 cutII.jpg|620px]] | ||
| - | |||
| + | [[File:SporeCoat.png|100px|right|link=Team:LMU-Munich/Spore_Coat_Proteins]] | ||
===Cloning Strategy=== | ===Cloning Strategy=== | ||
| - | + | <br> | |
<p align="justify">First, we used [http://partsregistry.org/Part:BBa_K823039 ''gfp''] as a proof of principle and fused it to ''cgeA'' and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823031 ''cotZ'']. This way, we can determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation.</p> | <p align="justify">First, we used [http://partsregistry.org/Part:BBa_K823039 ''gfp''] as a proof of principle and fused it to ''cgeA'' and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823031 ''cotZ'']. This way, we can determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation.</p> | ||
| Line 32: | Line 33: | ||
<p align="justify">Moreover, we wondered if clean deletions of the native spore crust genes would show any difference in fusion protein expression in our '''Sporo'''beads. Thus, we deleted the native ''cotZ'' and ''cgeA'' using the pMAD based gene deletion strategy described by [http://www.ncbi.nlm.nih.gov/pubmedterm=New%20Vector%20for%20Efficient%20Allelic%20Replacement%20in%20Naturally%20Nontransformable%2C%20Low-GC-Content%2C%20Gram-Positive%20Bacteria Arnaud ''et al''., 2004].</p> | <p align="justify">Moreover, we wondered if clean deletions of the native spore crust genes would show any difference in fusion protein expression in our '''Sporo'''beads. Thus, we deleted the native ''cotZ'' and ''cgeA'' using the pMAD based gene deletion strategy described by [http://www.ncbi.nlm.nih.gov/pubmedterm=New%20Vector%20for%20Efficient%20Allelic%20Replacement%20in%20Naturally%20Nontransformable%2C%20Low-GC-Content%2C%20Gram-Positive%20Bacteria Arnaud ''et al''., 2004].</p> | ||
| + | <br> | ||
| + | <p align="justify">In the end, we were able to finish five constructs and integrated them into wild type W168 and the Δ''cotZ'' mutant: | ||
| + | </p> | ||
| + | {| class="colored" width="100%" align="center" style="text-align:center;" | ||
| + | |- | ||
| + | |style="width:52%;"| | ||
| + | |style="width:20%;"| | ||
| + | |style="width:28%;"| | ||
| + | |- | ||
| + | ! | ||
| + | !recipient strain W168 | ||
| + | !recipient strain B 49 (W168 Δ''cotZ'') | ||
| + | |- | ||
| + | !pSB<sub>''Bs''</sub>1C-P<sub>''cotYZ''</sub>-''cotZ''-2aa-''gfp''-terminator | ||
| + | |B 53 | ||
| + | |B 70 | ||
| + | |- | ||
| + | !pSB<sub>''Bs''</sub>1C-P<sub>''cotYZ''</sub>-''cotZ''-''gfp''-terminator | ||
| + | |B 54 | ||
| + | |B 71 | ||
| + | |- | ||
| + | !pSB<sub>''Bs''</sub>1C-P<sub>''cotV''</sub>-''cotZ''-2aa-terminator | ||
| + | |B 55 | ||
| + | |B 72 | ||
| + | |- | ||
| + | ! pSB<sub>''Bs''</sub>1C-P<sub>''cotV''</sub>-''cotZ''-terminator | ||
| + | |B 56 | ||
| + | |B 73 | ||
| + | |- | ||
| + | !pSB<sub>''Bs''</sub>1C-P<sub>''cgeA''</sub>-''cotZ''-2aa-terminator | ||
| + | |B 52 | ||
| + | |B 69 | ||
| + | |- | ||
| + | |} | ||
| Line 39: | Line 74: | ||
| - | |||
| + | |||
| + | {| width="100%" cellpadding="20" | ||
| + | |[[File:LMU Arrow purple BACK.png|right|80px|link=Team:LMU-Munich/Spore_Coat_Proteins]] | ||
| + | |[[File:LMU Arrow purple NEXT.png|left|80px|link=Team:LMU-Munich/Spore_Coat_Proteins/result]] | ||
| + | |} | ||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Latest revision as of 13:17, 26 October 2012

The LMU-Munich team is exuberantly happy about the great success at the World Championship Jamboree in Boston. Our project Beadzillus finished 4th and won the prize for the "Best Wiki" (with Slovenia) and "Best New Application Project".
[ more news ]

Cloning Strategy
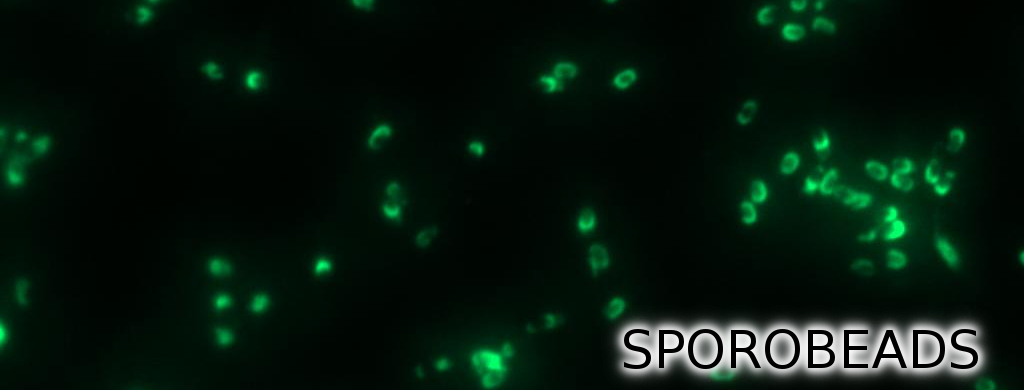
First, we used [http://partsregistry.org/Part:BBa_K823039 gfp] as a proof of principle and fused it to cgeA and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K823031 cotZ]. This way, we can determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation.
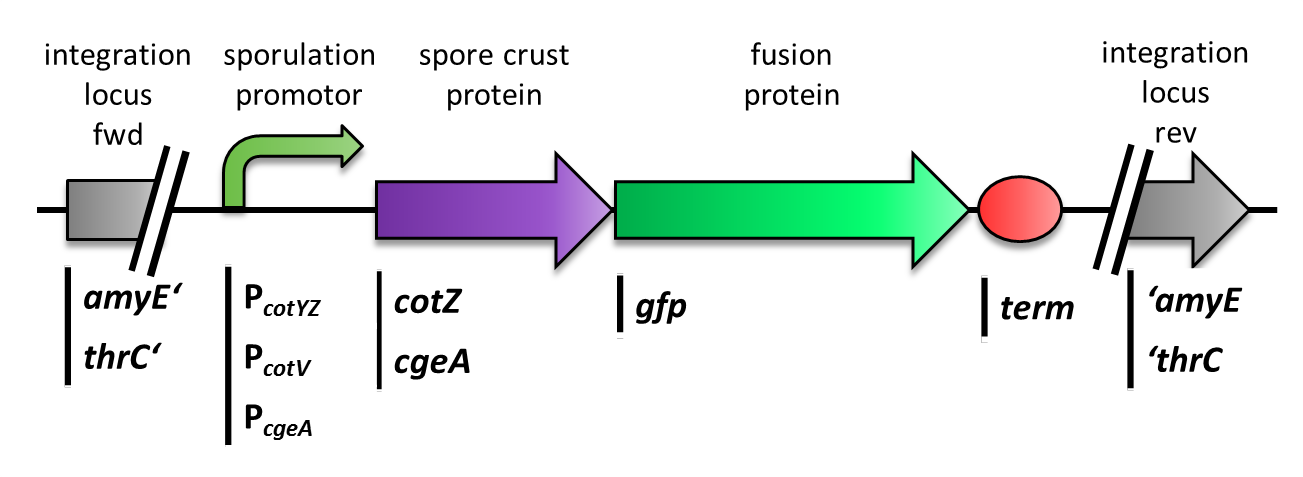
We constructed the BioBricks for cotZ, cgeA and gfp in [http://partsregistry.org/Help:Assembly_standard_25 Freiburg Standard]. The cotZ gene was then fused to its two native promoters, PcotV and to PcotYZ, and PcgeA, which regulates the transcription of cgeA. For cgeA we only used its native promoter PcgeA and the stronger one of the two promoters of the cotVWXYZ cluster, PcotYZ (for more details see crust promotor evaluation). In addition [http://partsregistry.org/Part:BBa_K823039 gfp] was ligated to the [http://partsregistry.org/wiki/index.php?title=Part:BBa_B0014 terminator B0014]. After sequence confirmation, we fused the cgeA/cotZ- and gfp-constructs together, applying the [http://partsregistry.org/Help:Assembly_standard_25 Freiburg standard] to create in-frame fusion proteins. This way, we created C-terminal gfp fusion to both spore crust proteins flanked by the promoters and terminator above (Fig. 4).
|
| |
|
But as we did not know if C- or N-terminal fusion would influence the fusion protein expression, our second aim was to construct N-terminal fusion proteins as well. For this purpose we wanted to fuse the genes for the crust proteins cotZ and cgeA to the terminator and gfp to the three chosen promoters. Unfortunately, there occured a mutation in the XbaI site during construction of gfp in Freiburg Standard. Therefore we were not able to finish these constructs. Finally we needed to clone our constructs into an empty Bacillus vector, so that they could get integrated into the genome of B. subtilis after transformation. For that purpose, we picked the empty vector pSBBS1C from our BacillusBioBrickBox, for the cotZ constructs. This vector integrates into the amyE locus, which allows to easily check the integration by starch test. In order to also express both crust protein constructs in one strain, the cgeA fusion proteins had to be cloned into another empty vector, pSBBS4S. Unfortunately, for unknown reasons, the cloning of the constructs with cgeA into this vector has so far not been successful.
Moreover, we wondered if clean deletions of the native spore crust genes would show any difference in fusion protein expression in our Sporobeads. Thus, we deleted the native cotZ and cgeA using the pMAD based gene deletion strategy described by [http://www.ncbi.nlm.nih.gov/pubmedterm=New%20Vector%20for%20Efficient%20Allelic%20Replacement%20in%20Naturally%20Nontransformable%2C%20Low-GC-Content%2C%20Gram-Positive%20Bacteria Arnaud et al., 2004].
In the end, we were able to finish five constructs and integrated them into wild type W168 and the ΔcotZ mutant:
| recipient strain W168 | recipient strain B 49 (W168 ΔcotZ) | |
|---|---|---|
| pSBBs1C-PcotYZ-cotZ-2aa-gfp-terminator | B 53 | B 70 |
| pSBBs1C-PcotYZ-cotZ-gfp-terminator | B 54 | B 71 |
| pSBBs1C-PcotV-cotZ-2aa-terminator | B 55 | B 72 |
| pSBBs1C-PcotV-cotZ-terminator | B 56 | B 73 |
| pSBBs1C-PcgeA-cotZ-2aa-terminator | B 52 | B 69 |
 "
"