|
|
| (3 intermediate revisions not shown) |
| Line 284: |
Line 284: |
| | <div class="container"> | | <div class="container"> |
| | <div id="access"> | | <div id="access"> |
| - | <div class="menu"><ul><li class="page_item page-item-264 current_page_item"><a href="http://uscigem2012.wordpress.com/background/">Background</a></li><li class="page_item page-item-2"><a href="https://2012.igem.org/Team:USC">E. Musici</a></li><li class="page_item page-item-5"><a href="https://2012.igem.org/Team:USC/Team">Team</a></li><li class="page_item page-item-10"><a href="https://2012.igem.org/Team:USC/Project">Project</a></li><li class="page_item page-item-12"><a href="https://2012.igem.org/Team:USC/Parts">Our BioBricks</a></li><li class="page_item page-item-14"><a href="https://2012.igem.org/Team:USC/Notebook">Lab Notebook</a></li><li class="page_item page-item-16"><a href="https://2012.igem.org/Team:USC/HumanPractices">Human Practices</a></li><li class="page_item page-item-45"><a href="https://2012.igem.org/Team:USC/Safety">Safety</a></li></ul></div> | + | <div class="menu"><ul><li class="page_item page-item-2"><a href="https://2012.igem.org/Team:USC">E. Musici</a></li><li class="page_item page-item-5"><a href="https://2012.igem.org/Team:USC/Team">Team</a></li><li class="page_item page-item-10 current_page_item"><a href="https://2012.igem.org/Team:USC/Project">Project</a></li><li class="page_item page-item-12"><a href=" |
| | + | https://2012.igem.org/Team:USC/Parts">Our BioBricks</a></li><li class="page_item page-item-14"><a href="https://2012.igem.org/Team:USC/Notebook">Lab Notebook</a></li><li class="page_item page-item-16"><a href="https://2012.igem.org/Team:USC/HumanPractices">Human Practices</a></li><li class="page_item page-item-45"><a href=" |
| | + | https://2012.igem.org/Team:USC/Safety">Safety</a></li></ul></div> |
| | </div><!-- #access --> | | </div><!-- #access --> |
| | </div><!-- .container --> | | </div><!-- .container --> |
| Line 311: |
Line 313: |
| | | | |
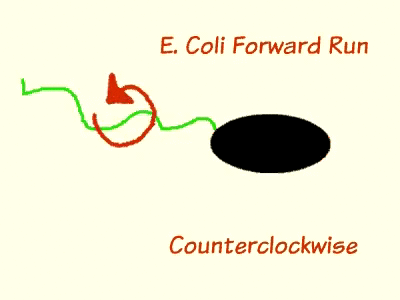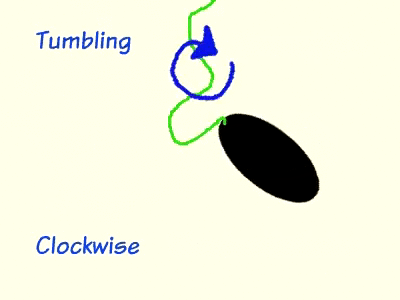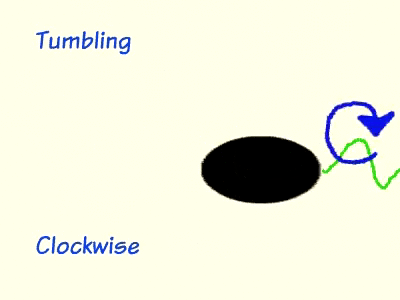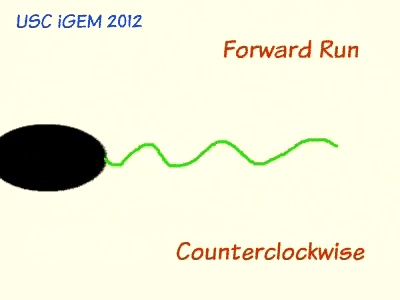
| | <p><span style="color:#000000;"><em>Escherichia coli</em> are able to move and change direction due to their locomotive sensory organelle, the flagella. When moving in an environment that lacks a concentration gradient, <em>E. coli</em> travels in one direction, stops, tumbles, and then continues moving in a new, random direction. In order for the cell to propel in a direction, the flagella form a tight bundle and spin counter-clockwise. After a brief period, the flagella reverse in rotation, inducing tumbling. When a concentration gradient is present in the media, bacteria have longer or shorter runs dependent on the chemical nature of the gradient, whether it is created by an attractant or a repellent. This is the result of chemotaxis, the phenomenon whereby organisms detect chemical compounds in their environment and alter behavior accordingly.<a href="http://uscigem2012.files.wordpress.com/2012/07/biobrick-gene-pathway.gif"><span style="color:#000000;"><img class="alignright size-medium wp-image-204" title="BioBrick-Gene-Pathway" src="http://uscigem2012.files.wordpress.com/2012/07/biobrick-gene-pathway.gif?w=300&h=300" alt="" width="300" height="300" /></span></a></span></p> | | <p><span style="color:#000000;"><em>Escherichia coli</em> are able to move and change direction due to their locomotive sensory organelle, the flagella. When moving in an environment that lacks a concentration gradient, <em>E. coli</em> travels in one direction, stops, tumbles, and then continues moving in a new, random direction. In order for the cell to propel in a direction, the flagella form a tight bundle and spin counter-clockwise. After a brief period, the flagella reverse in rotation, inducing tumbling. When a concentration gradient is present in the media, bacteria have longer or shorter runs dependent on the chemical nature of the gradient, whether it is created by an attractant or a repellent. This is the result of chemotaxis, the phenomenon whereby organisms detect chemical compounds in their environment and alter behavior accordingly.<a href="http://uscigem2012.files.wordpress.com/2012/07/biobrick-gene-pathway.gif"><span style="color:#000000;"><img class="alignright size-medium wp-image-204" title="BioBrick-Gene-Pathway" src="http://uscigem2012.files.wordpress.com/2012/07/biobrick-gene-pathway.gif?w=300&h=300" alt="" width="300" height="300" /></span></a></span></p> |
| - | <p><span style="color:#000000;">During chemotaxis, attractant chemicals bind to the cell membrane protein methyl-accepting chemotaxis receptors (MCP), which is bound to cheW and cheA on the cytoplasmic side of the cell membrane. cheW is an adaptor protein that serves as communication bridge between MCP and cheA. When MCP is unoccupied by an attractant, cheA is stimulated to self-phosphorylate using ATP and then donate the phosphate group to cheY. Phosphorylated cheY moves to the flagellum and induces clockwise rotation, which leads to tumbling. The phosphate group on cheY is cyclically removed by cheZ so that the bacterium remains responsive to changes in chemical concentration. As long as MCP remains unbound to an attractant, the bacterium will continue run and tumble pattern until it detects a change in the environment. When MCP is bound to a chemo attractant, the level of phosphorylated cheY drops significantly due to the inhibition of cheA self-phosphorylation from MCP. Flagella that are not attached to a phosphorylated cheY will spin in a counter-clockwise rotation and, resulting in a longer run by the bacteria.</span></p> | + | <p><span style="color:#000000;">During chemotaxis, attractant chemicals bind to the cell membrane protein methyl-accepting chemotaxis receptors (MCP), which is bound to <em>cheW</em> and <em>cheA</em> on the cytoplasmic side of the cell membrane. <em>cheW</em> is an adaptor protein that serves as communication bridge between MCP and <em>cheA</em>. When MCP is unoccupied by an attractant, <em>cheA</em> is stimulated to self-phosphorylate using ATP and then donate the phosphate group to <em>cheY</em>. Phosphorylated <em>cheY</em> moves to the flagellum and induces clockwise rotation, which leads to tumbling. The phosphate group on <em>cheY</em> is cyclically removed by <em>cheZ</em> so that the bacterium remains responsive to changes in chemical concentration. As long as MCP remains unbound to an attractant, the bacterium will continue run and tumble pattern until it detects a change in the environment. When MCP is bound to a chemo attractant, the level of phosphorylated <em>cheY</em> drops significantly due to the inhibition of <em>cheA</em> self-phosphorylation from MCP. Flagella that are not attached to a phosphorylated <em>cheY</em> will spin in a counter-clockwise rotation and, resulting in a longer run by the bacteria.</span></p> |
| - | <p><span style="color:#000000;">Our team aimed to uncouple the requirement for a chemo attractant and repellant from this system. To accomplish this we have generated BioBricks for the flagella apparatus as well as the chemotaxis signaling system for conditional expression. We have exploited this well-established regulatory mechanism as the foundation of our project. By controlling the expression of cheY and cheZ we can alter the rate of linear movement as well as tumbling frequency. By controlling the expression of all the flagella components we can effectively reconstitute the entire system in the <em>E. coli</em> B background, which lacks the flagella loci.</span></p> | + | <p><span style="color:#000000;">Our team aimed to uncouple the requirement for a chemo attractant and repellant from this system. To accomplish this we have generated BioBricks for the flagella apparatus as well as the chemotaxis signaling system for conditional expression. We have exploited this well-established regulatory mechanism as the foundation of our project. By controlling the expression of <em>cheY</em> and <em>cheZ</em> we can alter the rate of linear movement as well as tumbling frequency. By controlling the expression of all the flagella components we can effectively reconstitute the entire system in the <em>E. coli</em> B background, which lacks the flagella loci.</span></p> |
| | <p><a href="http://uscigem2012.files.wordpress.com/2012/07/singing_bacteria3.gif"><img class="aligncenter size-full wp-image-259" title="Singing_Bacteria" src="http://uscigem2012.files.wordpress.com/2012/07/bacteria_one_flagella3.gif?w=960" alt="" /></a></p> | | <p><a href="http://uscigem2012.files.wordpress.com/2012/07/singing_bacteria3.gif"><img class="aligncenter size-full wp-image-259" title="Singing_Bacteria" src="http://uscigem2012.files.wordpress.com/2012/07/bacteria_one_flagella3.gif?w=960" alt="" /></a></p> |
| | <p> </p> | | <p> </p> |
Background
Escherichia coli are able to move and change direction due to their locomotive sensory organelle, the flagella. When moving in an environment that lacks a concentration gradient, E. coli travels in one direction, stops, tumbles, and then continues moving in a new, random direction. In order for the cell to propel in a direction, the flagella form a tight bundle and spin counter-clockwise. After a brief period, the flagella reverse in rotation, inducing tumbling. When a concentration gradient is present in the media, bacteria have longer or shorter runs dependent on the chemical nature of the gradient, whether it is created by an attractant or a repellent. This is the result of chemotaxis, the phenomenon whereby organisms detect chemical compounds in their environment and alter behavior accordingly.
During chemotaxis, attractant chemicals bind to the cell membrane protein methyl-accepting chemotaxis receptors (MCP), which is bound to cheW and cheA on the cytoplasmic side of the cell membrane. cheW is an adaptor protein that serves as communication bridge between MCP and cheA. When MCP is unoccupied by an attractant, cheA is stimulated to self-phosphorylate using ATP and then donate the phosphate group to cheY. Phosphorylated cheY moves to the flagellum and induces clockwise rotation, which leads to tumbling. The phosphate group on cheY is cyclically removed by cheZ so that the bacterium remains responsive to changes in chemical concentration. As long as MCP remains unbound to an attractant, the bacterium will continue run and tumble pattern until it detects a change in the environment. When MCP is bound to a chemo attractant, the level of phosphorylated cheY drops significantly due to the inhibition of cheA self-phosphorylation from MCP. Flagella that are not attached to a phosphorylated cheY will spin in a counter-clockwise rotation and, resulting in a longer run by the bacteria.
Our team aimed to uncouple the requirement for a chemo attractant and repellant from this system. To accomplish this we have generated BioBricks for the flagella apparatus as well as the chemotaxis signaling system for conditional expression. We have exploited this well-established regulatory mechanism as the foundation of our project. By controlling the expression of cheY and cheZ we can alter the rate of linear movement as well as tumbling frequency. By controlling the expression of all the flagella components we can effectively reconstitute the entire system in the E. coli B background, which lacks the flagella loci.

 "
"
